library(sf)Linking to GEOS 3.10.2, GDAL 3.4.1, PROJ 8.2.1; sf_use_s2() is TRUElibrary(stars)Loading required package: abindlibrary(terra)terra 1.6.7Part 2: Extracting to buffers
We load sf and stars, and also terra which will be needed for extracting raster values to polygons.
library(sf)Linking to GEOS 3.10.2, GDAL 3.4.1, PROJ 8.2.1; sf_use_s2() is TRUElibrary(stars)Loading required package: abindlibrary(terra)terra 1.6.7Importing the points:
pnt = st_read('data/export/Address coordinates.gpkg')Reading layer `extracted' from data source
`/home/michael/Sync/presentations/p_2023_01_R_exposure_tutorial/data/export/Address coordinates.gpkg'
using driver `GPKG'
Simple feature collection with 13710 features and 2 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: 669825 ymin: 6578850 xmax: 695023 ymax: 6590699
Projected CRS: SWEREF99 TMpntSimple feature collection with 13710 features and 2 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: 669825 ymin: 6578850 xmax: 695023 ymax: 6590699
Projected CRS: SWEREF99 TM
First 10 features:
X_sw99_corr_final Y_sw99_corr_final geom
1 6579476 672969 POINT (672969 6579476)
2 6579535 672998 POINT (672998 6579535)
3 6580754 673589 POINT (673589 6580754)
4 6580685 670730 POINT (670730 6580685)
5 6580685 670730 POINT (670730 6580685)
6 6580707 670681 POINT (670681 6580707)
7 6580685 670730 POINT (670730 6580685)
8 6580063 671015 POINT (671015 6580063)
9 6581347 672437 POINT (672437 6581347)
10 6582407 673401 POINT (673401 6582407)plot(st_geometry(pnt))
Importing the raster:
r = read_stars('data/export/Greenness/NDVI/WholeSwedenNAv2017.tif')
names(r) = 'ndvi'
rstars object with 2 dimensions and 1 attribute
attribute(s):
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
ndvi 64 507 628 572.7616 695 855 151241
dimension(s):
from to offset delta refsys point values x/y
x 1 1012 669800 25 SWEREF99 TM FALSE NULL [x]
y 1 476 6590700 -25 SWEREF99 TM FALSE NULL [y]plot(r)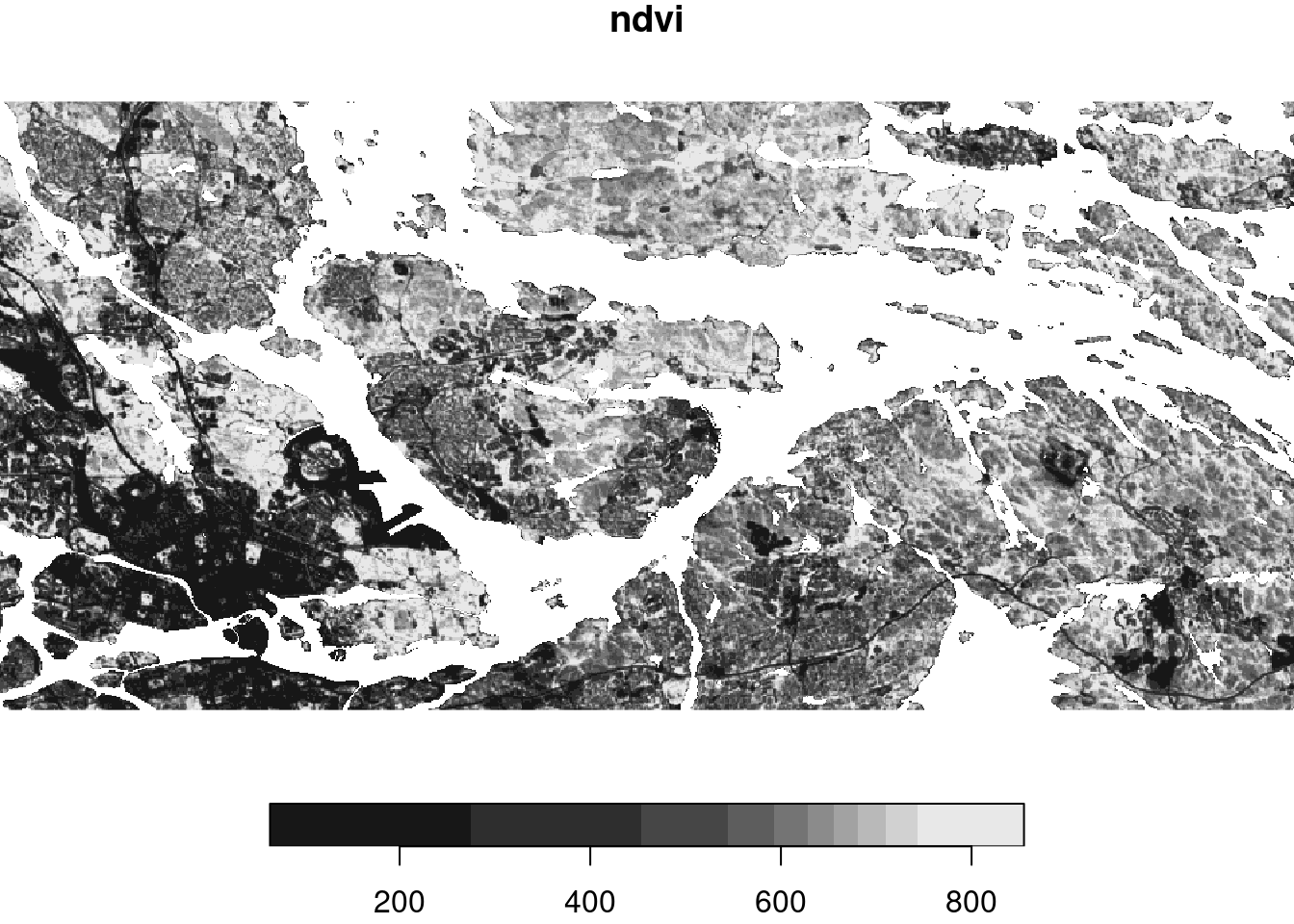
The st_buffer function is used to create buffers, with specified width (in CRS units). For example, here is how we can create 100, 250, and 500 \(m\) buffer layers, named pol_100, pol_250, and pol_500, respectively:
pol_100 = st_buffer(pnt, 100)
pol_250 = st_buffer(pnt, 250)
pol_500 = st_buffer(pnt, 500)Confirming that the CRS units are indeed \(m\) can be done as follows:
st_crs(pnt)$units[1] "m"Each of the layers pol_100, pol_250, and pol_500 is identical to the input pnt, only that the POINT geometries were replaced with POLYGON geometries of the respective buffers. For example:
pol_500Simple feature collection with 13710 features and 2 fields
Geometry type: POLYGON
Dimension: XY
Bounding box: xmin: 669325 ymin: 6578350 xmax: 695523 ymax: 6591199
Projected CRS: SWEREF99 TM
First 10 features:
X_sw99_corr_final Y_sw99_corr_final geom
1 6579476 672969 POLYGON ((673469 6579476, 6...
2 6579535 672998 POLYGON ((673498 6579535, 6...
3 6580754 673589 POLYGON ((674089 6580754, 6...
4 6580685 670730 POLYGON ((671230 6580685, 6...
5 6580685 670730 POLYGON ((671230 6580685, 6...
6 6580707 670681 POLYGON ((671181 6580707, 6...
7 6580685 670730 POLYGON ((671230 6580685, 6...
8 6580063 671015 POLYGON ((671515 6580063, 6...
9 6581347 672437 POLYGON ((672937 6581347, 6...
10 6582407 673401 POLYGON ((673901 6582407, 6...Figure 3 shows the 500-\(m\) buffers:
plot(st_geometry(pol_500))
and Figure 4 shows all three (100, 250, and 500 \(m\)) buffers for one point:
plot(st_geometry(pnt[1, ]), pch = 3, extent = st_bbox(pol_500[1, ]))
plot(st_geometry(pol_500[1, ]), border = 'red', add = TRUE)
plot(st_geometry(pol_250[1, ]), border = 'green', add = TRUE)
plot(st_geometry(pol_100[1, ]), border = 'blue', add = TRUE)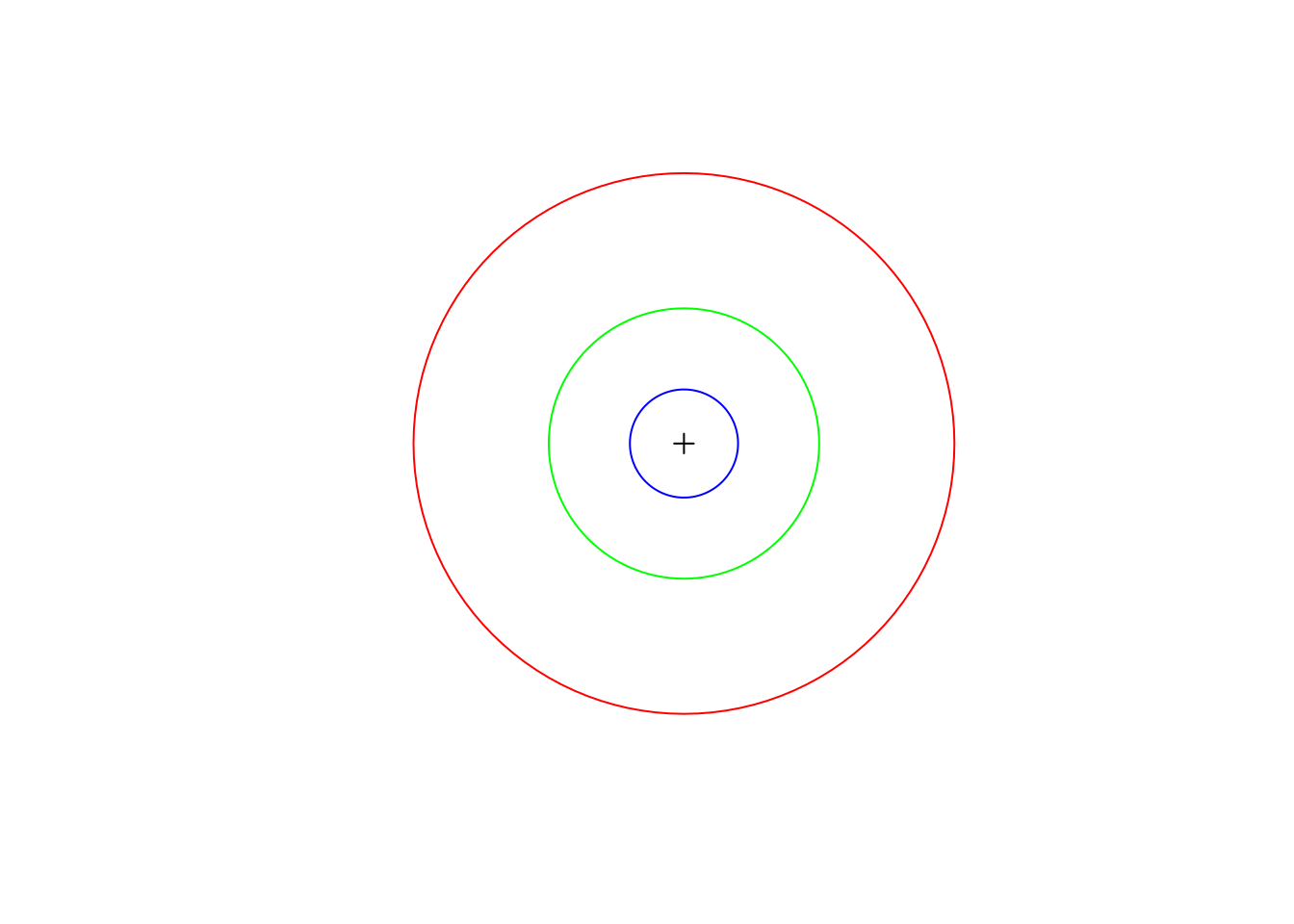
starsExtracting raster values to polygons in the sf+stars framework can be done using function aggregate, similarly to the base method for data.frame. For example, to calculate the median r value in each pol_100 buffer excluding NA, we use:
x = aggregate(r, pol_100, median, na.rm = TRUE)
x = st_as_sf(x)
xSimple feature collection with 13710 features and 1 field
Geometry type: POLYGON
Dimension: XY
Bounding box: xmin: 669725 ymin: 6578750 xmax: 695123 ymax: 6590799
Projected CRS: SWEREF99 TM
First 10 features:
ndvi geom
1 292.5 POLYGON ((673069 6579476, 6...
2 268.0 POLYGON ((673098 6579535, 6...
3 202.0 POLYGON ((673689 6580754, 6...
4 304.0 POLYGON ((670830 6580685, 6...
5 NA POLYGON ((670830 6580685, 6...
6 355.0 POLYGON ((670781 6580707, 6...
7 NA POLYGON ((670830 6580685, 6...
8 295.0 POLYGON ((671115 6580063, 6...
9 118.0 POLYGON ((672537 6581347, 6...
10 186.0 POLYGON ((673501 6582407, 6...However (!), probably for compatibility with other aggregate methods in R, each pixel can only be assigned into one grouping polygon. Therefore overlapping polygons capture the values of “unassigned” pixels only:
aggregate(r, pol_100[rep(1,3), ], median, na.rm = TRUE)[[1]][1] 292.5 NA NAThis means that aggregate works incorrectly for any polygon layer that has overlapping polygons, such as our buffer layers (Figure 3).
terraThe terra package provides another framework for working with rasters in R. There is very large overlap in functionality between stars and terra, with few exceptions. For example, terra has an extract function for summarizing raster values per polygons, without the above-mentioned issue that aggregate has.
The terra package has its own classes for rasters and vector layers. Before using terra::extract we therefore need to transform our objects to the terra classes:
stars→SpatRaster, using function rastsf→SpatVector, using function vectFor example:
pol_100_t = vect(pol_100)
pol_100_t class : SpatVector
geometry : polygons
dimensions : 13710, 2 (geometries, attributes)
extent : 669725, 695123, 6578750, 6590799 (xmin, xmax, ymin, ymax)
coord. ref. : SWEREF99 TM (EPSG:3006)
names : X_sw99_corr_final Y_sw99_corr_final
type : <int> <int>
values : 6579476 672969
6579535 672998
6580754 673589r_t = rast(r)
r_tclass : SpatRaster
dimensions : 476, 1012, 1 (nrow, ncol, nlyr)
resolution : 25, 25 (x, y)
extent : 669800, 695100, 6578800, 6590700 (xmin, xmax, ymin, ymax)
coord. ref. : SWEREF99 TM (EPSG:3006)
source : memory
name : lyr.1
min value : 64
max value : 855 Now we can run extract, using identical syntax as aggregate:
x = extract(r_t, pol_100_t, median, na.rm = TRUE)The resulting object is a data.frame with the polygon IDs and the summary of the attribute(s):
head(x) ID lyr.1
1 1 292.5
2 2 230.0
3 3 202.0
4 4 304.0
5 5 304.0
6 6 341.0We can attach the values to the point layer pnt as follows:
pnt$ndvi_100 = x$lyr.1
head(pnt)Simple feature collection with 6 features and 3 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: 670681 ymin: 6579476 xmax: 673589 ymax: 6580754
Projected CRS: SWEREF99 TM
X_sw99_corr_final Y_sw99_corr_final geom ndvi_100
1 6579476 672969 POINT (672969 6579476) 292.5
2 6579535 672998 POINT (672998 6579535) 230.0
3 6580754 673589 POINT (673589 6580754) 202.0
4 6580685 670730 POINT (670730 6580685) 304.0
5 6580685 670730 POINT (670730 6580685) 304.0
6 6580707 670681 POINT (670681 6580707) 341.0Here we do the same (in one step) also for pol_250 and pol_500, resulting in the attributes ndvi_250 and ndvi_500, respectively:
pnt$ndvi_250 = extract(r_t, vect(pol_250), median, na.rm = TRUE)$lyr.1
pnt$ndvi_500 = extract(r_t, vect(pol_500), median, na.rm = TRUE)$lyr.1
pntSimple feature collection with 13710 features and 5 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: 669825 ymin: 6578850 xmax: 695023 ymax: 6590699
Projected CRS: SWEREF99 TM
First 10 features:
X_sw99_corr_final Y_sw99_corr_final geom ndvi_100 ndvi_250
1 6579476 672969 POINT (672969 6579476) 292.5 406.0
2 6579535 672998 POINT (672998 6579535) 230.0 385.5
3 6580754 673589 POINT (673589 6580754) 202.0 210.0
4 6580685 670730 POINT (670730 6580685) 304.0 385.0
5 6580685 670730 POINT (670730 6580685) 304.0 385.0
6 6580707 670681 POINT (670681 6580707) 341.0 399.0
7 6580685 670730 POINT (670730 6580685) 304.0 385.0
8 6580063 671015 POINT (671015 6580063) 295.0 261.5
9 6581347 672437 POINT (672437 6581347) 118.0 122.0
10 6582407 673401 POINT (673401 6582407) 186.0 183.0
ndvi_500
1 360.0
2 362.0
3 110.0
4 420.0
5 420.0
6 428.0
7 420.0
8 365.0
9 206.5
10 198.0The median values extracted from the various buffers, for each point, are shown in Figure 5:
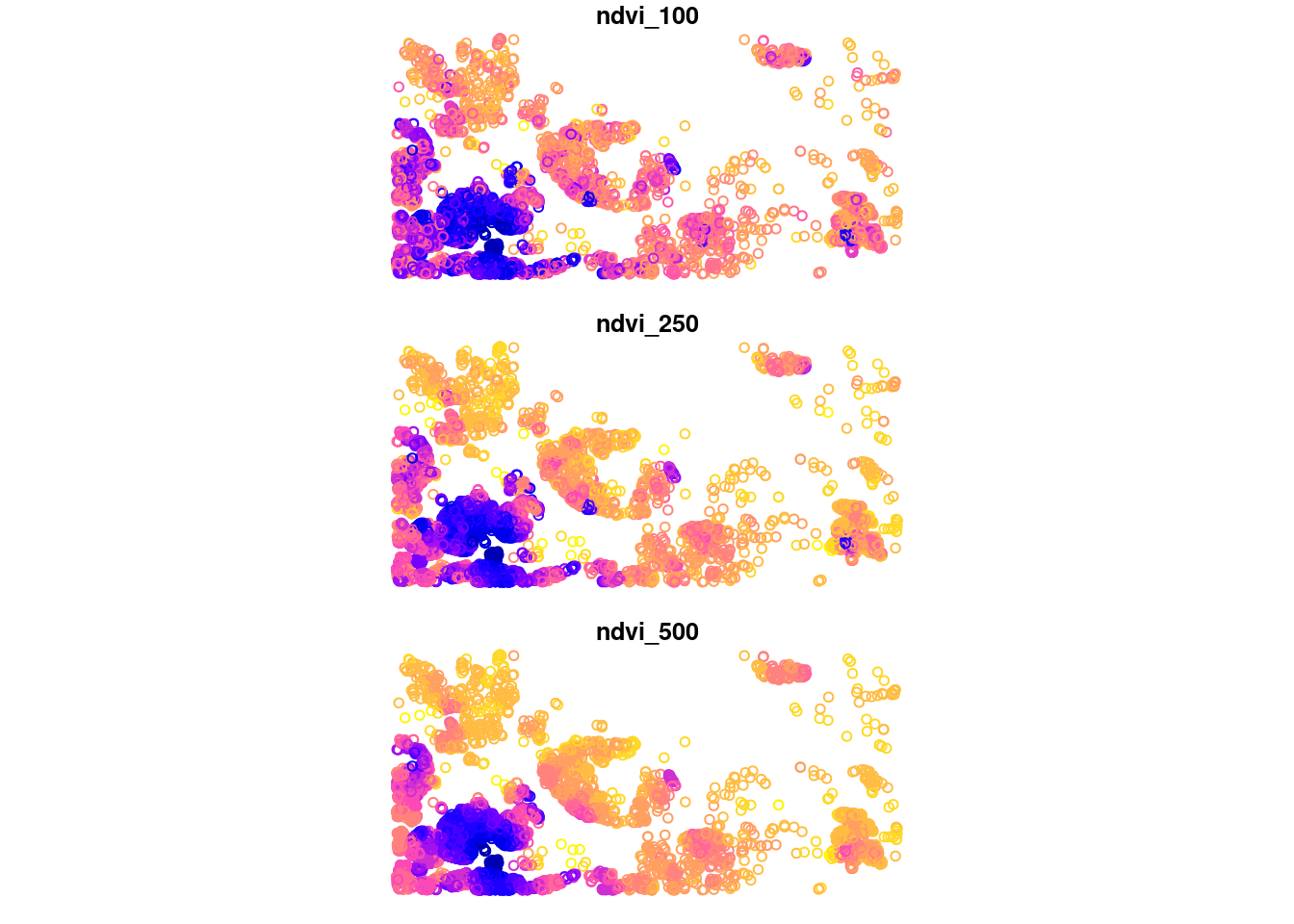
plot(pnt[c('ndvi_100', 'ndvi_250', 'ndvi_500')])