Chapter 6 Raster processing
Last updated: 2023-10-17 21:41:14
Aims
Our aims in this chapter are:
- Learn how to get raster subsets
- Learn to set raster dimension properties
- Learn to calculate new rasters, based on one or more overlapping rasters, using raster algebra
We will use the following R packages:
stars
6.1 Time series for single pixel
6.1.1 Selecting pixel by row & column
In most of the examples in this chapter, we are going to use the MOD13A3_2000_2019.tif raster which we already met in Section 5.3. Let’s read it into memory:
Keep in mind that this is a raster time series, where the “layers” refer to NDVI images taken in different times. As an exercise of accessing raster values (Section 5.3.8.4) and working with time series (Section 3.1.3), in this Section we are going to extract and plot a NDVI time series for a single pixel.
In Section 5.3.8.2, we saw how the matrix or array of raster values can be accessed using the [[ operator, as in r[[1]]. As another exercise, let’s access a “slice” of a single pixel through all of the raster layers, as follows:
The result is a vector (why?). Plotting the vector displays a time-series of NDVI at a particular location (Figure 6.1):
Figure 6.1: Single pixel values across all layers
Now, let us improve the time series plot, by:
6.1.2 Combining values with dates
The file MOD13A3_2000_2019_dates2.csv corresponds to the third dimension of MOD13A3_2000_2019.tif, containing the dates when each NDVI image was captured. This file includes the additional season column which we calculated in Section 4.6.
Using the MOD13A3_2000_2019_dates2.csv table, we can display dates on the x-axis (Figure 6.2). First we read the dates table:
Then we can plot the dates on the x-axis, as follows:
Figure 6.2: Single pixel values across all layers
6.1.3 Visualizing seasonal pattern
Figure 6.2 clearly shows the seasonal pattern of NDVI. However, we can further improve the interpretability of the time series plot if we mark seasons with different color. One way to do that is to plot the NDVI observations of each season one at a time, using a for loop.
As a first step, we define the vector of seasons (seasons) and the corresponding vector of colors (cols):
Then, we can use a for loop (Section 4.2.3) to mark the portion of the NDVI time series from each season (Figure 6.3):
plot(dates$date, v, type = 'l', xlab = 'Time', ylab = 'NDVI', col = 'grey')
for(i in 1:length(seasons)) {
tmp = v
tmp[dates$season != seasons[i]] = NA
lines(dates$date, tmp, col = cols[i], type = 'o')
}Figure 6.3: Single pixel values across all layers
6.2 Raster subsetting
6.2.1 Selecting rows, column and layers
We can get a subset of a stars object while keeping all of its properties, using the [ operator.
The stars subset operator [ works as follows:
- The 1st index selects attributes
- The 2nd index selects the first dimension, typically
[x], i.e., raster columns - The 3rd index selects the second dimension, typically
[y], i.e., raster rows - The 4th index selects the third dimension (if there is one), typically
band, i.e., raster layers - (Subsequent indices, if any, select the remaining dimensions)
In this book, as well as in most spatial data analysis practices, we encounter two types of rasters: single-band rasters and multi-band rasters (Section 5.3.2). Accordingly, typical subsetting of stars objects requires either 3 or 4 indices, where the first index is left blank (since there is only one attribute). Again (Section 5.3.8.4), keep in mind that the order of stars indices is opposite of the order we are used to in R, since the first dimension refers to columns [x], while the second dimension refers to rows [y].
For example, the following expression creates a subset with two columns, three rows and four layers from r:
s = r[, 100:101, 200:202, 2:5]
s
## stars object with 3 dimensions and 1 attribute
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## NDVI 0.1575 0.1841 0.2261 0.2235417 0.2696 0.2852
## dimension(s):
## from to offset delta refsys point x/y
## x 100 101 3239946 926.6 unnamed FALSE [x]
## y 200 202 3717158 -926.6 unnamed FALSE [y]
## band 2 5 NA NA NA NAThe raster subset is plotted in Figure 6.4:
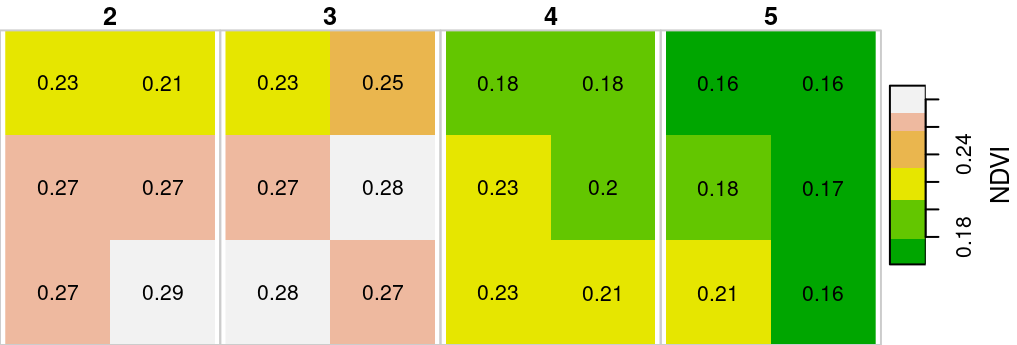
Figure 6.4: Raster subset with two columns, three rows and four layers
Note that we use the round function to round all raster values to two decimal places, so that the text labels can fit inside the pixels. This is an example of a raster algebra operation, which we learn about later in this chapter (Section 6.4).
What is the meaning of the following plot (Figure 6.5)?
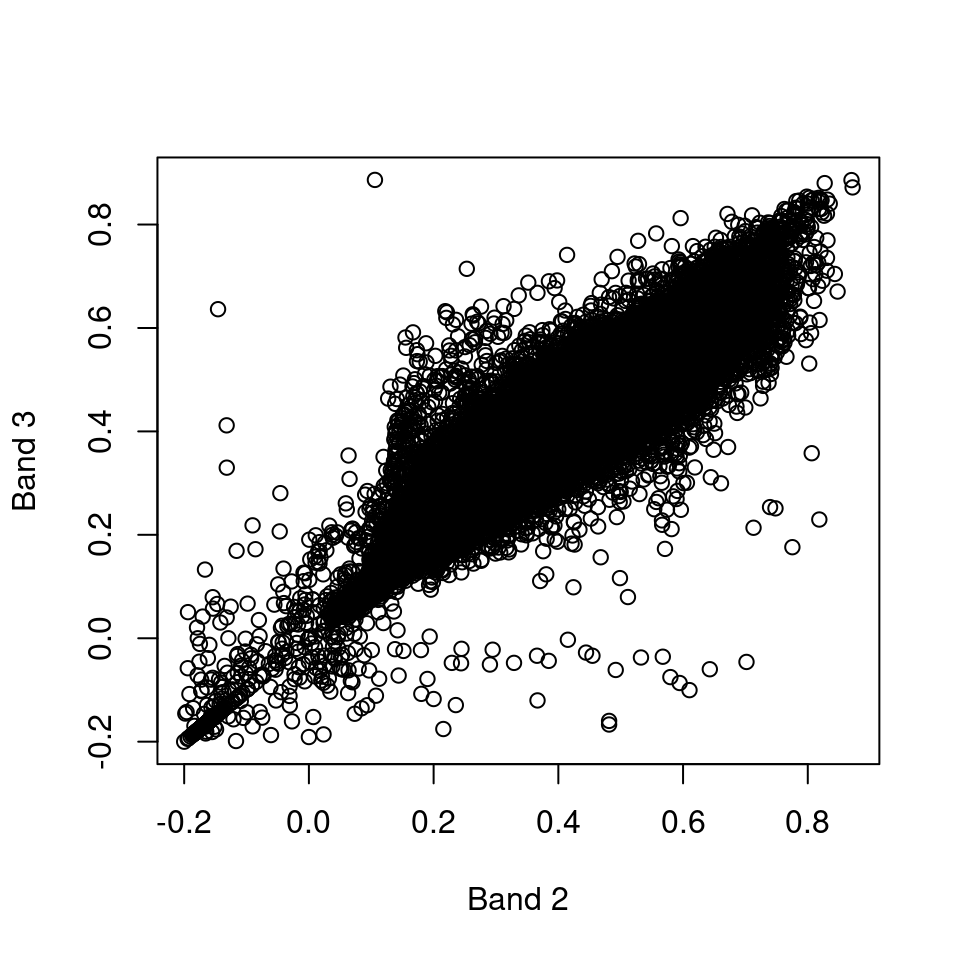
Figure 6.5: Band 3 as function of band 2
6.2.2 Dropping unnecessary dimensions
When a stars subset includes just one step along the 3rd dimension (in general: any dimension other than x and y), the “unnecessary” demension can be dropped using drop=TRUE. For example, compare the output of the following two expressions. Both expressions return a subset consisting of the 7th layer of r. However, the first expression returns a three-dimensional stars object (with a single layer in the 3rd dimension):
r[,,,7]
## stars object with 3 dimensions and 1 attribute
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## NDVI -0.1986 0.0959 0.1117 0.1482469 0.1887 0.8518 9877
## dimension(s):
## from to offset delta refsys point x/y
## x 1 167 3239946 926.6 unnamed FALSE [x]
## y 1 485 3717158 -926.6 unnamed FALSE [y]
## band 7 7 NA NA NA NAwhile the second expression returns a two-dimensional stars object, where the 3rd dimension was “dropped”:
r[,,,7, drop = TRUE]
## stars object with 2 dimensions and 1 attribute
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## NDVI -0.1986 0.0959 0.1117 0.1482469 0.1887 0.8518 9877
## dimension(s):
## from to offset delta refsys point x/y
## x 1 167 3239946 926.6 unnamed FALSE [x]
## y 1 485 3717158 -926.6 unnamed FALSE [y]Accordingly, the raster values are represented as an array in the first case, and as a matrix in the second case:
Note that the meaning of the drop parameter in stars subsetting differs from what we learned about in data.frame (Section 4.1.5.3), matrix (Section 5.1.7.1), and array (Section 5.2.2) subsetting, in two respects. First, in stars subsetting, drop=TRUE specifies dropping of raster dimensions, while in data.frame, matrix, and array subsetting, drop specifies simplifying to a different data structure (a vector or a matrix). Second, the default of drop in stars is drop=FALSE; in data.frame and matrix subsetting, the default is drop=TRUE.
6.3 Raster dimensions
6.3.1 Getting dimension properties
As mentioned in Section 5.3.8.3, the st_dimensions function can be used to access dimension properties of a stars object. The function resurns an object of class dimensions. Printing the object gives all information on the raster dimensions in the form of a table:
st_dimensions(r)
## from to offset delta refsys point x/y
## x 1 167 3239946 926.6 unnamed FALSE [x]
## y 1 485 3717158 -926.6 unnamed FALSE [y]
## band 1 233 NA NA NA NAInternally, the dimensions object is a list of dimensions, with one list element per dimension. Each element is also a list, with the internal list elements being the various properties of a single dimension:
str(st_dimensions(r))
## List of 3
## $ x :List of 7
## ..$ from : num 1
## ..$ to : num 167
## ..$ offset: num 3239946
## ..$ delta : num 927
## ..$ refsys:List of 2
## .. ..$ input: chr "unnamed"
## .. ..$ wkt : chr "PROJCRS[\"unnamed\",\n BASEGEOGCRS[\"unnamed ellipse\",\n DATUM[\"unknown\",\n ELLIPSOID[\"| __truncated__
## .. ..- attr(*, "class")= chr "crs"
## ..$ point : logi FALSE
## ..$ values: NULL
## ..- attr(*, "class")= chr "dimension"
## $ y :List of 7
## ..$ from : num 1
## ..$ to : num 485
## ..$ offset: num 3717158
## ..$ delta : num -927
## ..$ refsys:List of 2
## .. ..$ input: chr "unnamed"
## .. ..$ wkt : chr "PROJCRS[\"unnamed\",\n BASEGEOGCRS[\"unnamed ellipse\",\n DATUM[\"unknown\",\n ELLIPSOID[\"| __truncated__
## .. ..- attr(*, "class")= chr "crs"
## ..$ point : logi FALSE
## ..$ values: NULL
## ..- attr(*, "class")= chr "dimension"
## $ band:List of 7
## ..$ from : num 1
## ..$ to : int 233
## ..$ offset: num NA
## ..$ delta : num NA
## ..$ refsys: chr NA
## ..$ point : logi NA
## ..$ values: NULL
## ..- attr(*, "class")= chr "dimension"
## - attr(*, "raster")=List of 4
## ..$ affine : num [1:2] 0 0
## ..$ dimensions : chr [1:2] "x" "y"
## ..$ curvilinear: logi FALSE
## ..$ blocksizes : int [1:233, 1:2] 167 167 167 167 167 167 167 167 167 167 ...
## .. ..- attr(*, "dimnames")=List of 2
## .. .. ..$ : NULL
## .. .. ..$ : chr [1:2] "x" "y"
## ..- attr(*, "class")= chr "stars_raster"
## - attr(*, "class")= chr "dimensions"Individual dimensions can be accessed using the $ operator by list element name, or the [[ operator by list element index or name (more on lists in Section 11.1). For example, all three expressions shown below return the same result, the properties of the 1st dimension 'x':
str(st_dimensions(r)[[1]])
## List of 7
## $ from : num 1
## $ to : num 167
## $ offset: num 3239946
## $ delta : num 927
## $ refsys:List of 2
## ..$ input: chr "unnamed"
## ..$ wkt : chr "PROJCRS[\"unnamed\",\n BASEGEOGCRS[\"unnamed ellipse\",\n DATUM[\"unknown\",\n ELLIPSOID[\"| __truncated__
## ..- attr(*, "class")= chr "crs"
## $ point : logi FALSE
## $ values: NULL
## - attr(*, "class")= chr "dimension"str(st_dimensions(r)[['x']])
## List of 7
## $ from : num 1
## $ to : num 167
## $ offset: num 3239946
## $ delta : num 927
## $ refsys:List of 2
## ..$ input: chr "unnamed"
## ..$ wkt : chr "PROJCRS[\"unnamed\",\n BASEGEOGCRS[\"unnamed ellipse\",\n DATUM[\"unknown\",\n ELLIPSOID[\"| __truncated__
## ..- attr(*, "class")= chr "crs"
## $ point : logi FALSE
## $ values: NULL
## - attr(*, "class")= chr "dimension"str(st_dimensions(r)$x)
## List of 7
## $ from : num 1
## $ to : num 167
## $ offset: num 3239946
## $ delta : num 927
## $ refsys:List of 2
## ..$ input: chr "unnamed"
## ..$ wkt : chr "PROJCRS[\"unnamed\",\n BASEGEOGCRS[\"unnamed ellipse\",\n DATUM[\"unknown\",\n ELLIPSOID[\"| __truncated__
## ..- attr(*, "class")= chr "crs"
## $ point : logi FALSE
## $ values: NULL
## - attr(*, "class")= chr "dimension"Properties of individual dimensions can be accessed through further subsetting (Figure 6.6). For example, the following expression returns the offset (origin) and delta (resolution) of the 'x' dimension of raster r:
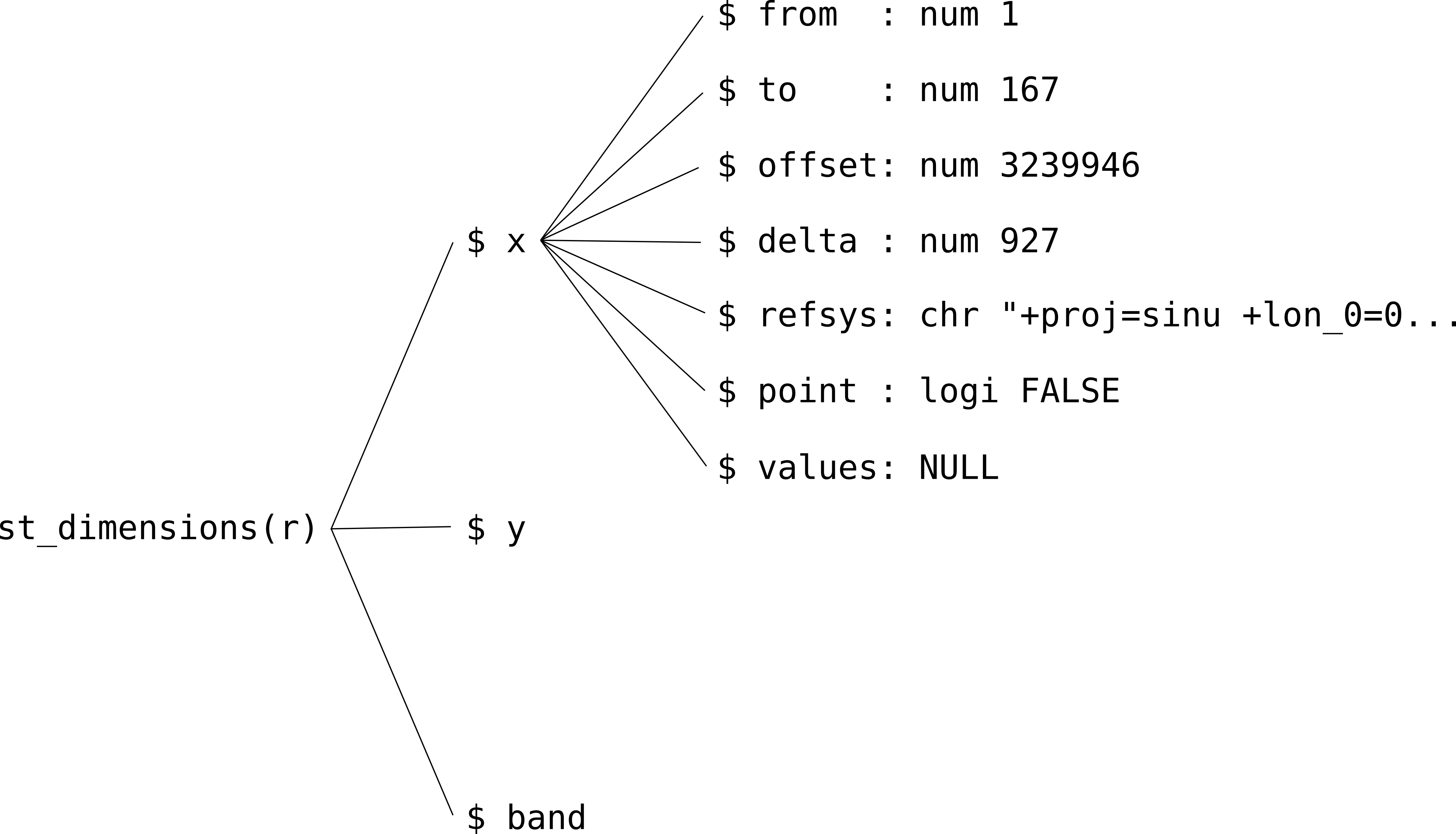
Figure 6.6: The hierarchical structure of stars dimensions properties
The meanings of the seven properties that every stars dimension has are summarized in Table 6.1.
| Property | Contents | Description |
|---|---|---|
from |
numeric length 1 |
The start index |
to |
numeric length 1 |
The end index |
offset |
numeric length 1 |
The start coordinate (or time) of the first element (e.g., the pixel/cell boundary) |
delta |
numeric length 1 |
The increment / cell size (e.g., the resolution) |
refsys |
character, or crs |
The Coordinate Reference System (e.g., the CRS) |
point |
logical length 1 |
Whether cells/pixels refer to areas/periods (FALSE), or to points/instances (TRUE) |
values |
NULL, or an object that gives the specific dimension values |
The dimension values, in case they are not calculated from offset and delta |
For more information on stars dimensions, see the official stars Data Model article.
6.3.2 Setting dimension properties
A .tif file cannot hold metadata regarding raster “layers”, such as the date when the images were captured in case the layers comprise a time series, or the band names in case the layers comprise spectral bands (Section 6.4.2). Accordingly, the third dimension of a stars object created from GeoTIFF file does not contain any information other than the from and to indices (e.g., 1 and 233):
st_dimensions(r)
## from to offset delta refsys point x/y
## x 1 167 3239946 926.6 unnamed FALSE [x]
## y 1 485 3717158 -926.6 unnamed FALSE [y]
## band 1 233 NA NA NA NAWe can incorporate more information into a stars object dimensions, or modify the existing information, on our own. This is done using function st_set_dimensions, where we need to specify:
.x—Thestarsobjectwhich—The index (e.g.,1) or name (e.g.,'x') of the dimension to be modified
as well as the dimension properties and their new values, such as:
offset—The offset (i.e., origin)delta—The delta (i.e., resolution)23values—The values along the dimensionnames—The new dimension names
as in st_set_dimensions(r, 'x', offset=0, delta=1000).
Additionally, the names of all dimensions can be modified using (i.e., without specifying which):
.x— Thestarsobjectnames—The new dimension names
as in st_set_dimensions(r, names=c('a','b','c')).
For example, for a non-spatial dimension that represents time, such as the 'band' dimension in r, we can:
- set the values to the corresponding dates or time (such as
Datevalues), and - change the dimension name to
time,
as follows:
Now, the 3rd dimension of r is named 'time' and contains the Date values:
st_dimensions(r)
## from to offset delta refsys point values x/y
## x 1 167 3239946 926.6 unnamed FALSE NULL [x]
## y 1 485 3717158 -926.6 unnamed FALSE NULL [y]
## time 1 233 NA NA Date NA 2000-02-15,...,2019-06-15If necessary, the dimension values can be read back using st_get_dimension_values. For example:
z = st_get_dimension_values(r, 'time')
class(z)
## [1] "Date"
z[1:10]
## [1] "2000-02-15" "2000-03-15" "2000-04-15" "2000-05-15" "2000-06-15"
## [6] "2000-07-15" "2000-08-15" "2000-09-15" "2000-10-15" "2000-11-15"What is the meaning of the dimension values given by
st_get_dimension_values(r,'x')andst_get_dimension_values(r,'y')? Given thatst_dimensions(r)$y$valuesandst_dimensions(r)$y$valuesareNULL, where do you think these dimension values come from?
Why do we need to bother with editing or supplementing dimension properties? One reason is that it is more convenient to have all of the relevant raster data in the same object (rather than two separate objects, such as r and dates). Moreover, information regarding stars dimensions is utilized by stars functions in various contexts. For example, the layer values apprear as labels in plot output panels (Figure 6.7):
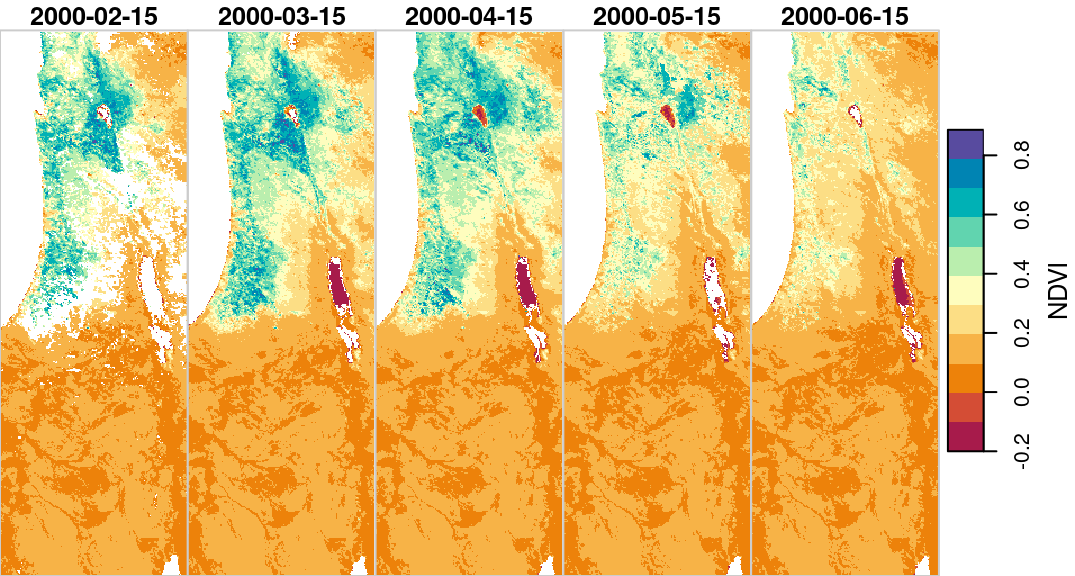
Figure 6.7: Date labels for raster layers, i.e., the third raster dimension
or as column values when converting from stars to data.frame:
6.3.3 Converting a matrix to raster
6.3.3.1 The volcano matrix
Converting a matrix to a stars raster is rarely needed in practice, since most of the time you will be working with existing rasters imported from files (Section 5.3.2). However, experimenting with the matrix→stars conversion will help us better understand the nature of rasters, in general, and of the stars class, in particular.
When converting a matrix or an array to a stars raster, we must manually set the properties of the dimensions, most importantly the spatial dimensions 'x' and 'y'.
In the first example, we will convert the built-in volcano matrix (Section 5.1.6) to a stars raster. First, we convert the transposed (why? see Section 5.3.8.4) matrix to a stars raster, using function st_as_stars:
v = st_as_stars(t(volcano))
v
## stars object with 2 dimensions and 1 attribute
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## A1 94 108 124 130.1879 150 195
## dimension(s):
## from to offset delta point x/y
## X1 1 61 0 1 FALSE [x]
## X2 1 87 0 1 FALSE [y]Then, we may want to set the dimension and attribute names:
v = st_set_dimensions(v, names = c('x', 'y'))
names(v) = 'elevation'
v
## stars object with 2 dimensions and 1 attribute
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## elevation 94 108 124 130.1879 150 195
## dimension(s):
## from to offset delta point x/y
## x 1 61 0 1 FALSE [x]
## y 1 87 0 1 FALSE [y]Finally, we set the offset and delta of the x and y dimensions, or, other words, where do these axes start from (i.e., origin), and what is their step size (i.e., resolution). The offset values of 0 (see below), in this case, is arbitrary. The delta is also arbitrary, but should be equal for both 'x' and 'y' dimensions (unless we want to have asymmetrical pixels). In this case, according to the ?volcano help page, the matrix represents a \(10 \times 10\) \(m\) grid. Accordingly, we use delta values of 10 and -10 so that the coordinate units are “meters”. The delta of the y-axis is negative (!), so that the values decrease when going to from 1st matrix column to last:
v = st_set_dimensions(v, 'x', offset = 0, delta = 10)
v = st_set_dimensions(v, 'y', offset = 0, delta = -10)
v
## stars object with 2 dimensions and 1 attribute
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## elevation 94 108 124 130.1879 150 195
## dimension(s):
## from to offset delta x/y
## x 1 61 0 10 [x]
## y 1 87 0 -10 [y]Compare the output of
plot(v,axes=TRUE)before and after setting theoffsetanddeltato see the effect of those settings.
The result is shown in Figure 6.8. Note that we need to use the reset=FALSE argument whenever we want to have additional layers, such as a contours, in a stars plot:
plot(v, axes = TRUE, breaks = 'equal', col = terrain.colors(100), key.pos = 4, reset = FALSE)
contour(v, add = TRUE)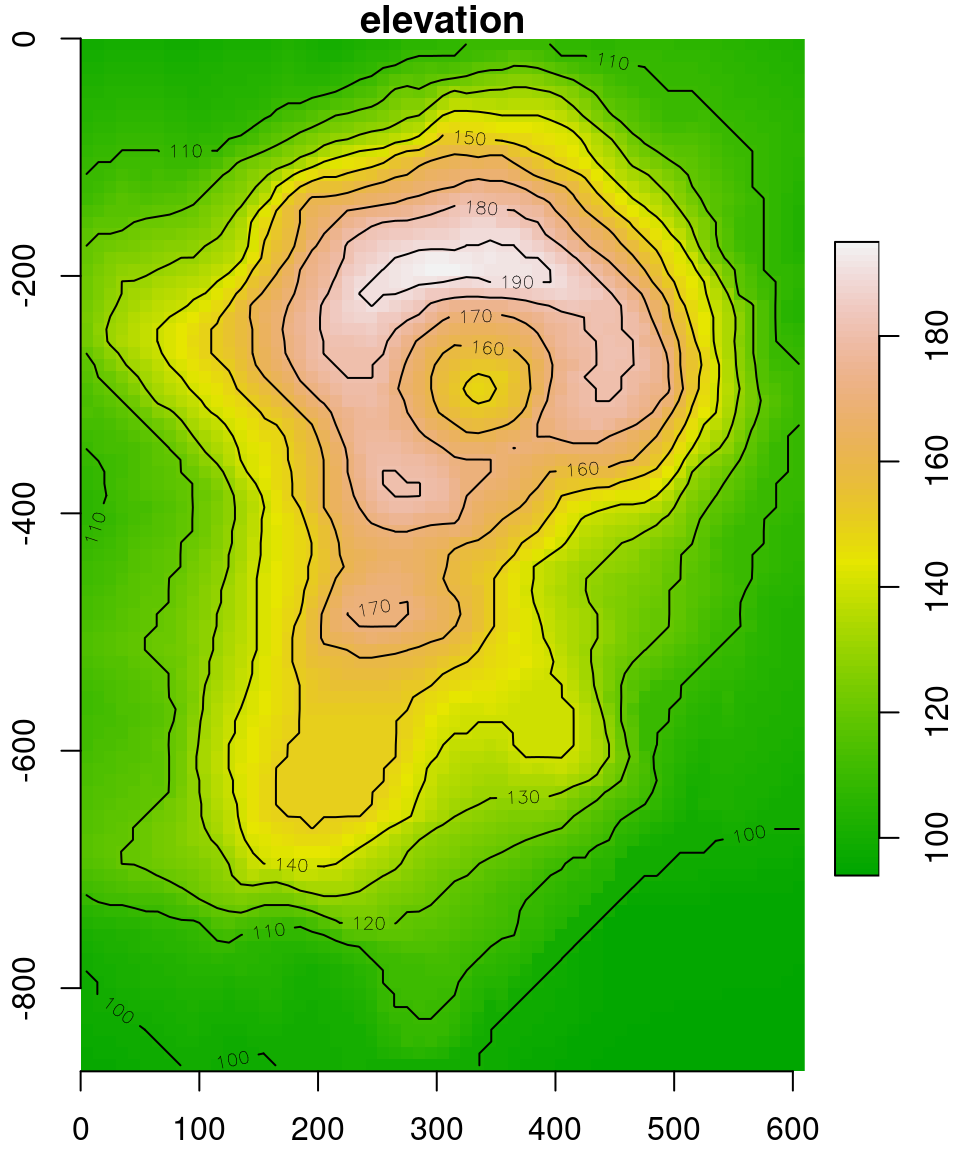
Figure 6.8: The volcano matrix converted to stars raster
The fact that the offset is arbitrary is demonstrated with the following expression, which “shifts” the raster so that the (0, 0) coordinate is in the botton-left rather than top-left corner:
The modified volcano raster is shown in Figure 6.9.
plot(v, axes = TRUE, breaks = 'equal', col = terrain.colors(100), key.pos = 4, reset = FALSE)
contour(v, add = TRUE)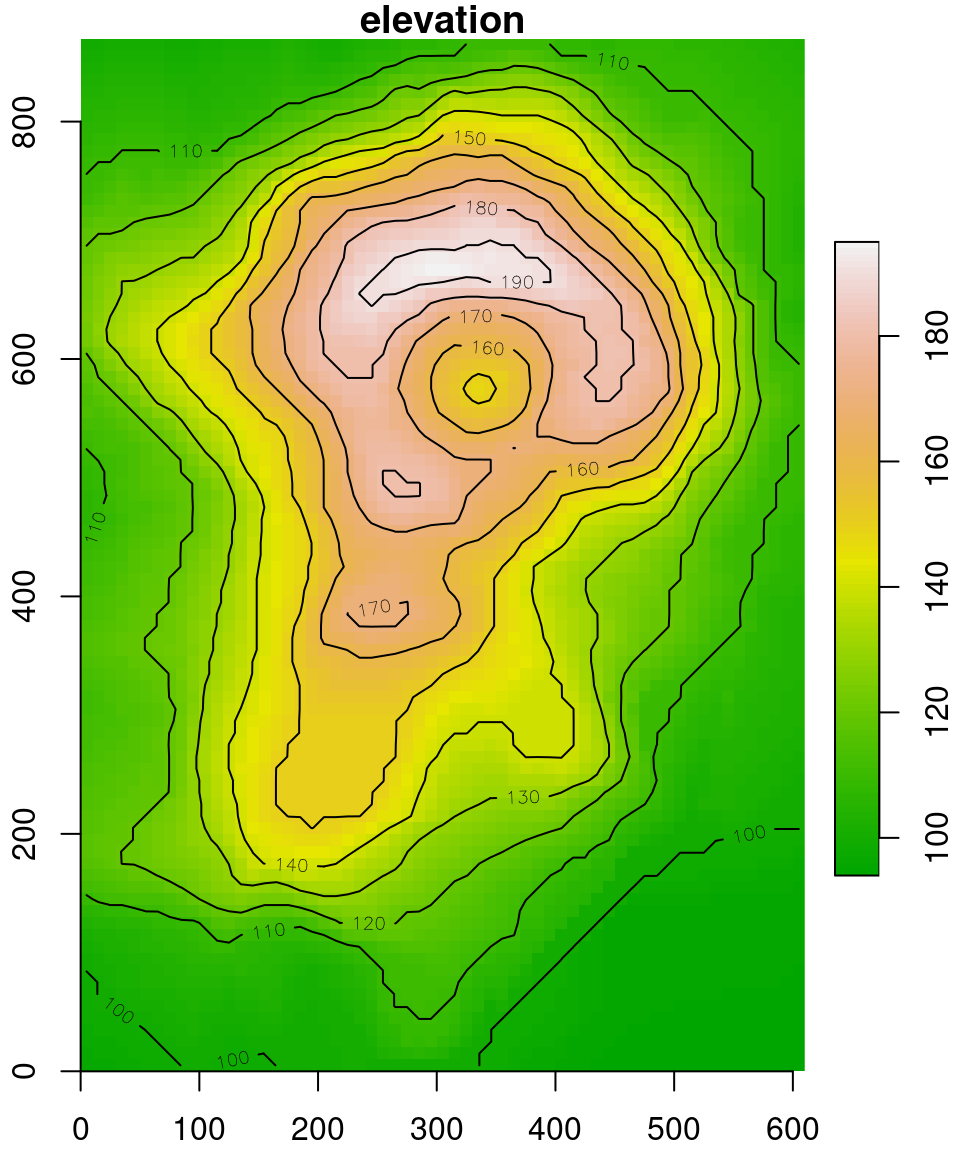
Figure 6.9: The volcano raster with modified offset
Comparing Figures 6.8 and 6.9, the only thing that has changed are the y-axis coordinates.
6.3.3.2 The Haifa DEM matrix
In the second example, we will recreate the dem.tif small DEM of Haifa (Figure 5.12), starting from a plain matrix. Here is the matrix we start with:
v = c(
NA, NA, NA, NA, NA, 3, 3,
NA, NA, NA, NA, 4, 6, 4,
NA, NA, NA, NA, 6, 9, 7,
NA, 61, 9, 4, 9, 10, 16,
NA, 106, 132, 11, 6, 6, 27,
NA, 47, 254, 146, 6, 6, 12,
NA, 31, 233, 340, 163, 13, 64,
3, 39, 253, 383, 448, 152, 39,
5, 32, 199, 357, 414, 360, 48,
7, 49, 179, 307, 403, 370, 55
)
m = matrix(v, nrow = 10, ncol = 7, byrow = TRUE)
m
## [,1] [,2] [,3] [,4] [,5] [,6] [,7]
## [1,] NA NA NA NA NA 3 3
## [2,] NA NA NA NA 4 6 4
## [3,] NA NA NA NA 6 9 7
## [4,] NA 61 9 4 9 10 16
## [5,] NA 106 132 11 6 6 27
## [6,] NA 47 254 146 6 6 12
## [7,] NA 31 233 340 163 13 64
## [8,] 3 39 253 383 448 152 39
## [9,] 5 32 199 357 414 360 48
## [10,] 7 49 179 307 403 370 55This time, assuming we know the right spatial reference information, we can create a realistically georeferenced stars object. To position a matrix or array in geographic space (Section 5.3.1), we need to know the coordinates of the starting point (offset) and the resolution (delta) for the 'x' and 'y' dimensions. We also need to know the CRS that the offset and delta refer to.
For example, the following code section sets the two offset values (679624 and 3644759), the resolution (2880) and the CRS (32636). Note that the CRS is specified using st_set_crs and an EPSG code. In this case, the EPSG code 32636 specifies the CRS named “UTM zone 36N”. Again, the y-axis resolution is negative, so that going along the matrix columns translates to decreasing y-axis values.
s = st_as_stars(t(m))
s = st_set_dimensions(s, names = c('x', 'y'))
s = st_set_dimensions(s, 'x', offset = 679624, delta = 2880)
s = st_set_dimensions(s, 'y', offset = 3644759, delta = -2880)
s = st_set_crs(s, 32636)
names(s) = 'elevation'The result is identical to dem.tif (Figure 5.14), as shown in Figure 6.10:
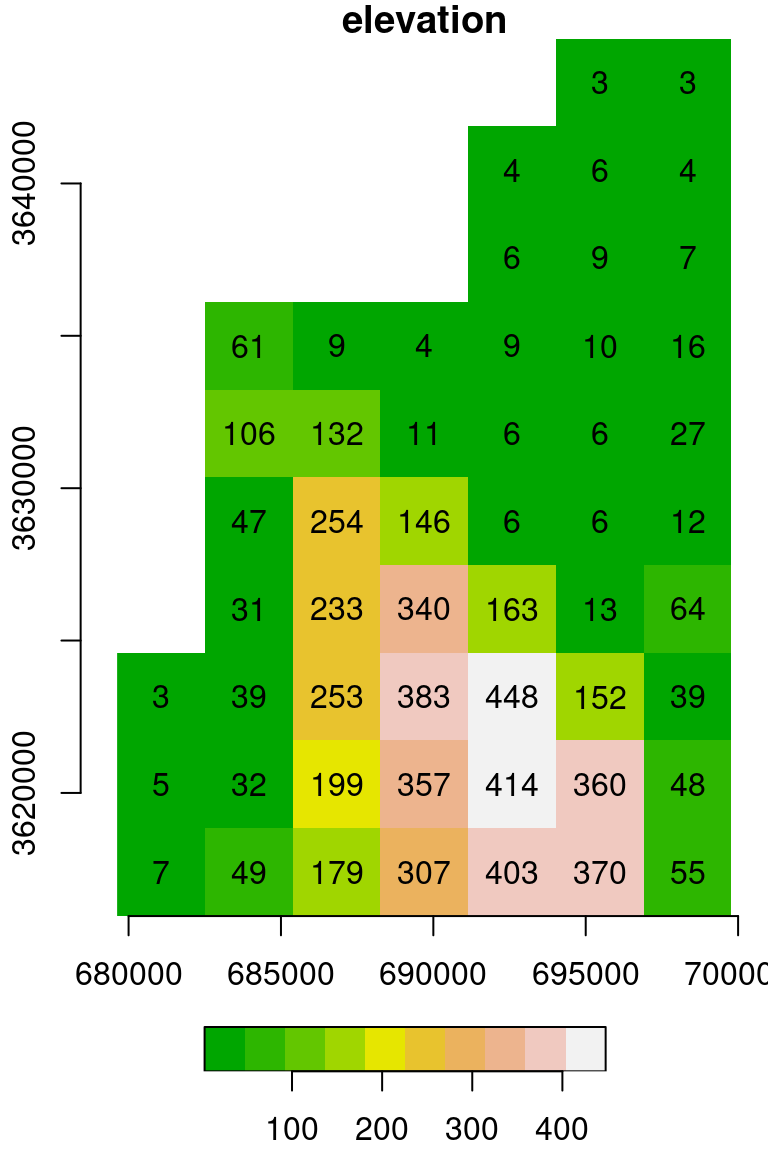
Figure 6.10: matrix converted to a stars object: Digital Elevation Model (DEM) of haifa
The effect of transposing the input matrix and modifying the sign of the y-axis delta is summarized in Figure 6.11. The figure demonstrates that only the scenario where delta_y is negative (while delta_x is positive) and the input matrix is transposed, ensure that the “print” orientation of the matrix matches the spatial orientation of the resulting raster.
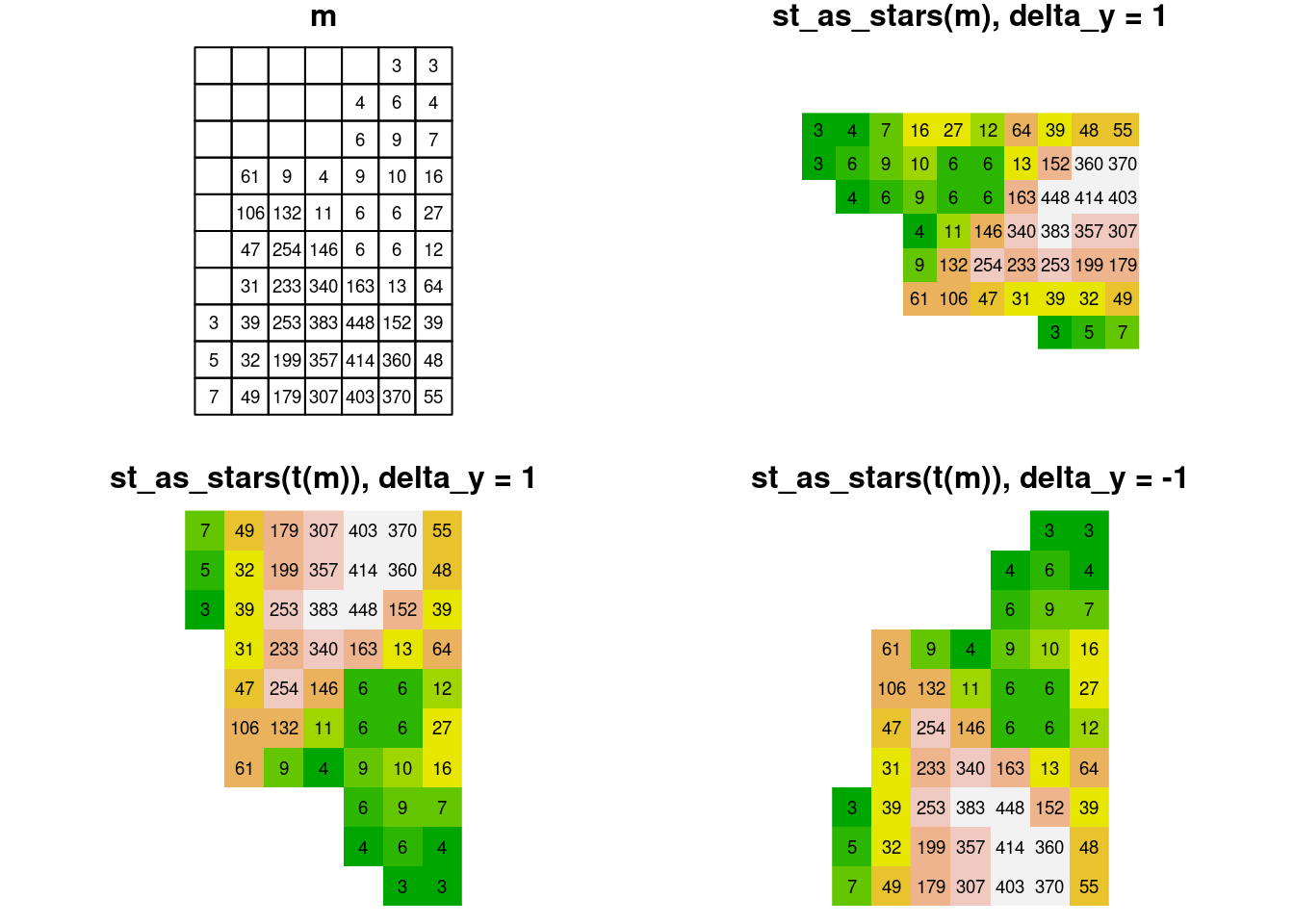
Figure 6.11: Converting matrix to stars: the effect of transposing the input matrix and modifying the sign of the y-axis delta
6.4 Raster algebra
6.4.1 Arithmetic and logical operations on layers
Raster algebra is the application of functions on a cell-by-cell basis, repeatedly for all pixels of one or more overlapping rasters. For example, we can apply the + function on two overlapping rasters to obtain a new raster, where each pixel value is the sum of the corresponding pixel values from the two inputs (Figure 6.12).
Figure 6.12: Raster algebra (http://rpubs.com/etiennebr/visualraster)
Similarly, we can use any arithmetic (Section 1.3.2) or conditional (Section 1.3.4) operator:
- Arithmetic:
+,-,*,/ - Logical:
<,<=,>,>=,==,!=,!
to make calculations on each pair of overlapping rasters, to get a new raster where each pixel value is the result of the given operation on the overlapping pixels in the input rasters. The operations can also combine rasters and numeric values, in which case the numeric value is recycled, as in r>5.
We can also use functions such as:
- Functions:
is.na,abs,round,ceiling,floor,sqrt,log,log10,exp,cos,sin
which are applied on an individual raster, transforming each of its pixels, separately.
For the next few examples, let’s create two single-band rasters named x and y. Note that we are using round to transform each of the rasters to rounded values with two decimal places (so that it is more convenient to display text labels):
x = r[, 99:101, 202:204, 1, drop = TRUE]
y = r[, 99:101, 202:204, 2, drop = TRUE]
x = round(x, 2)
y = round(y, 2)Figure 6.13 shows the two rasters x and y:
plot(x, text_values = TRUE, main = 'x', col = terrain.colors(5))
plot(y, text_values = TRUE, main = 'y', col = terrain.colors(5))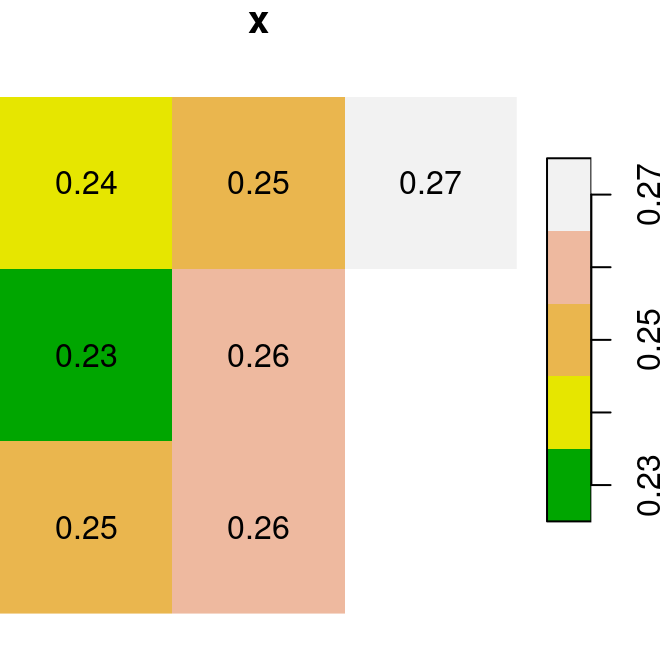
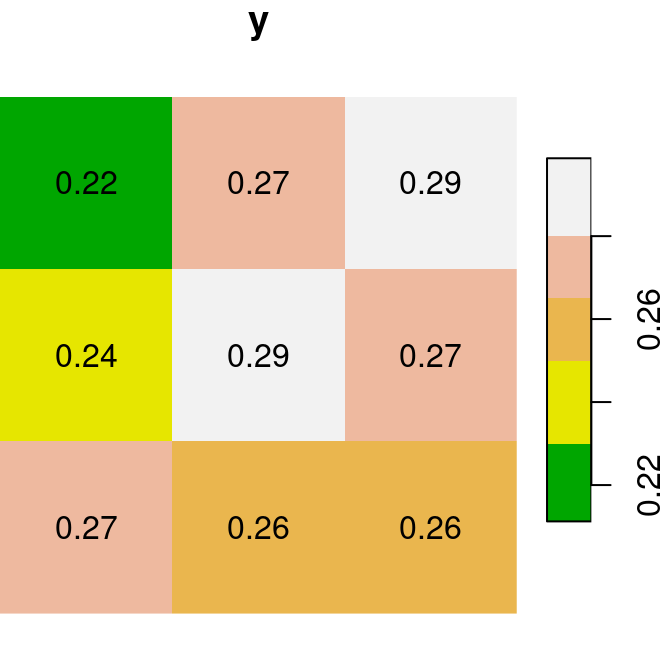
Figure 6.13: Two small stars rasters x and y to demonstrate raster algebra
As a first example of raster algebra, we calculate x+y, where each pixel value is the sum of two corresponding pixels in x and y (Figure 6.14):
plot(x, text_values = TRUE, main = 'x', col = terrain.colors(5))
plot(y, text_values = TRUE, main = 'y', col = terrain.colors(5))
plot(x + y, text_values = TRUE, main = 'x+y', col = terrain.colors(5))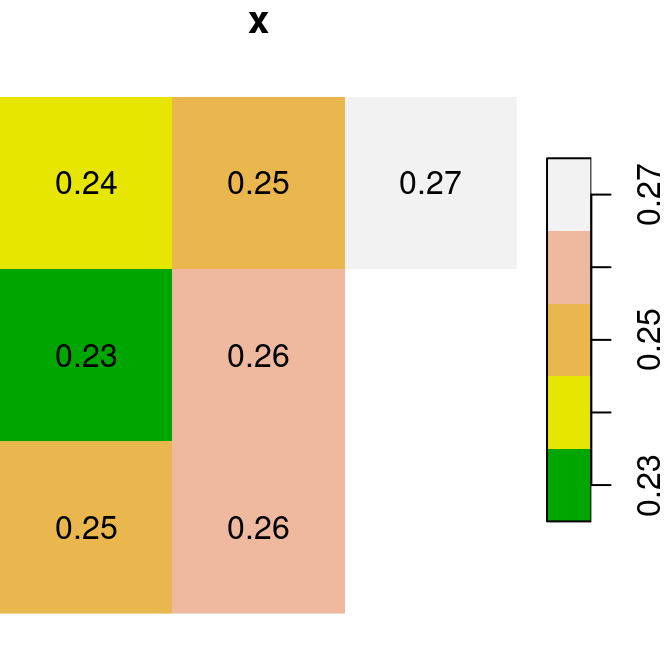
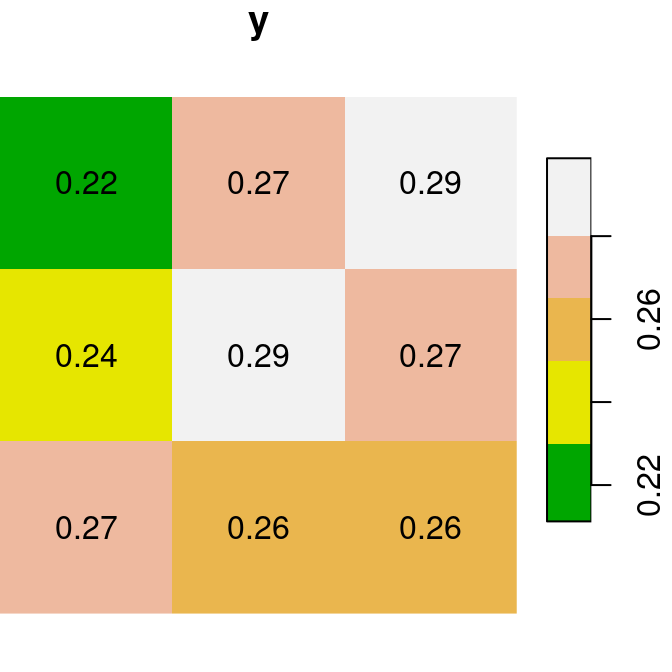
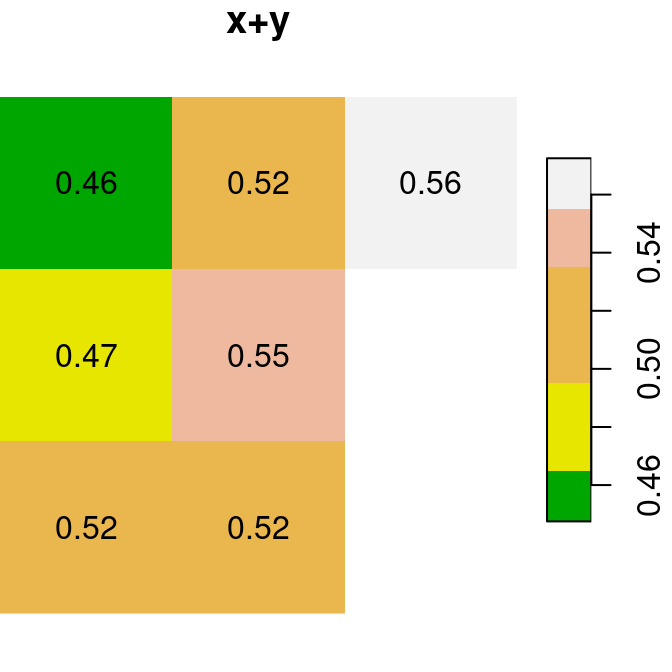
Figure 6.14: x, y and x+y
Why are two pixels in the bottom-right of
x+yin Figure 6.14 displayed without any labels?
Here are several other examples of raster algebra: x^2, x*y and x+5. Note that x^2 and x+5 combine a raster and a numeric value, rather than two rasters, in which case the numeric value is recycled (Figure 6.15):
plot(x^2, text_values = TRUE, main = 'x^2', col = terrain.colors(5))
plot(x * y, text_values = TRUE, main = 'x*y', col = terrain.colors(5))
plot(x + 5, text_values = TRUE, main = 'x+5', col = terrain.colors(5))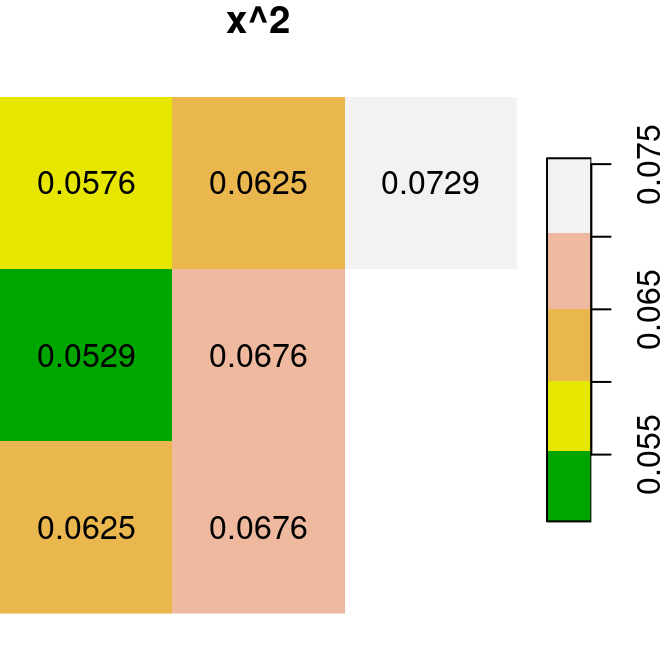
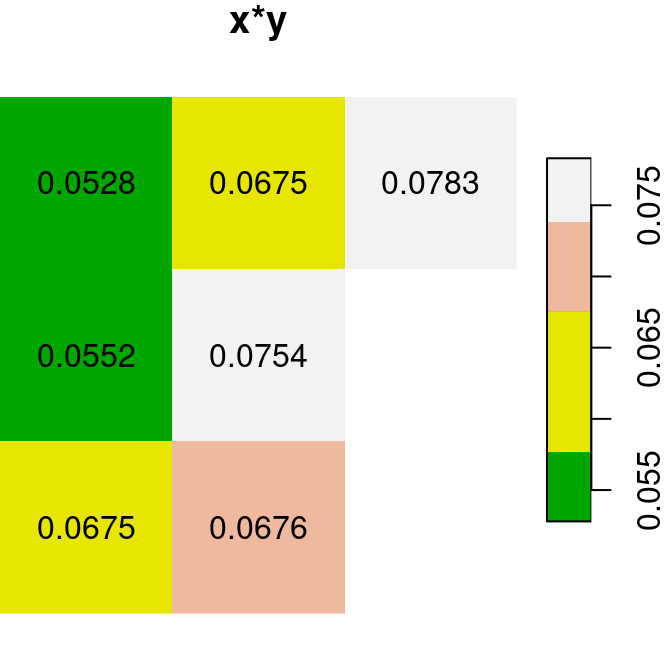
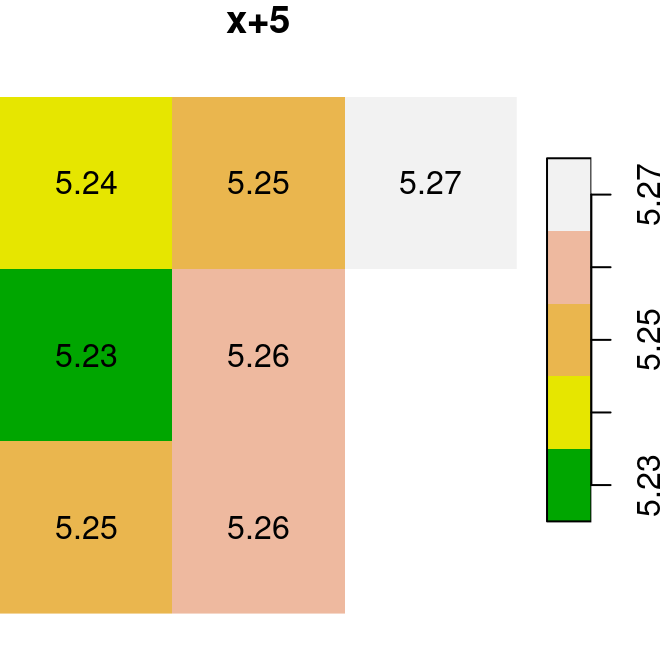
Figure 6.15: x^2, x*y and x+5
A logical raster algebra operation produces a logical raster, a raster where pixel values are TRUE or FALSE (or NA). For example, Figure 6.16 shows the logical rasters x>0.25, x<y and is.na(x):
plot(x > 0.25, text_values = TRUE, main = 'x>0.25', col = terrain.colors(2, rev = TRUE))
plot(x < y, text_values = TRUE, main = 'x<y', col = terrain.colors(2, rev = TRUE))
plot(is.na(x), text_values = TRUE, main = 'is.na(x)', col = terrain.colors(2, rev = TRUE))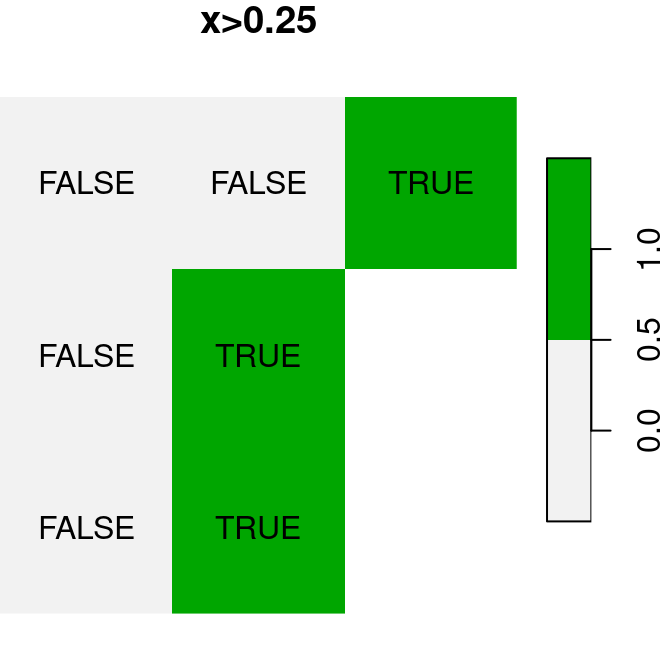
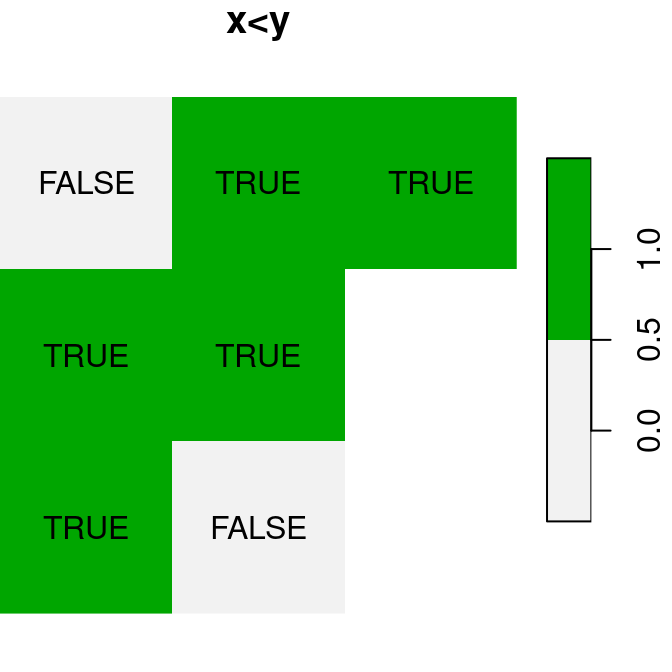
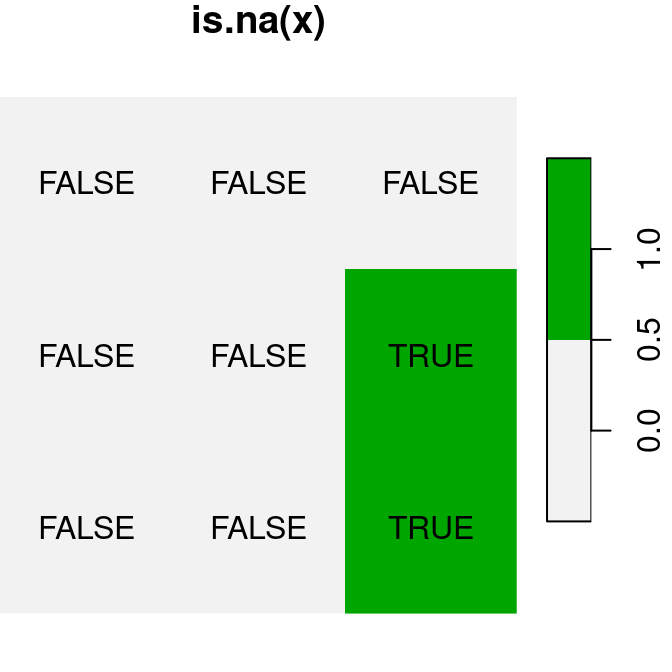
Figure 6.16: Logical raster algebra operations: x>0.25, x<y and is.na(x)
The last three examples produce logical rasters (Figure 6.16), which we have not encountered so far. In operations where a numeric representation is required, such as:
- An arithmetic operation
- Saving to a file
logical raster values TRUE and FALSE automatically become 1 and 0, respectively, similarly to what happens with vectors (Section 2.3.10.1). For example (Figure 6.17):
plot(is.na(x), text_values = TRUE, main = 'is.na(x)', col = terrain.colors(2, rev = TRUE))
plot(is.na(x) * 1, text_values = TRUE, main = 'is.na(x)*1', col = terrain.colors(2, rev = TRUE))
plot(is.na(x) + 30, text_values = TRUE, main = 'is.na(x)+30', col = terrain.colors(2, rev = TRUE))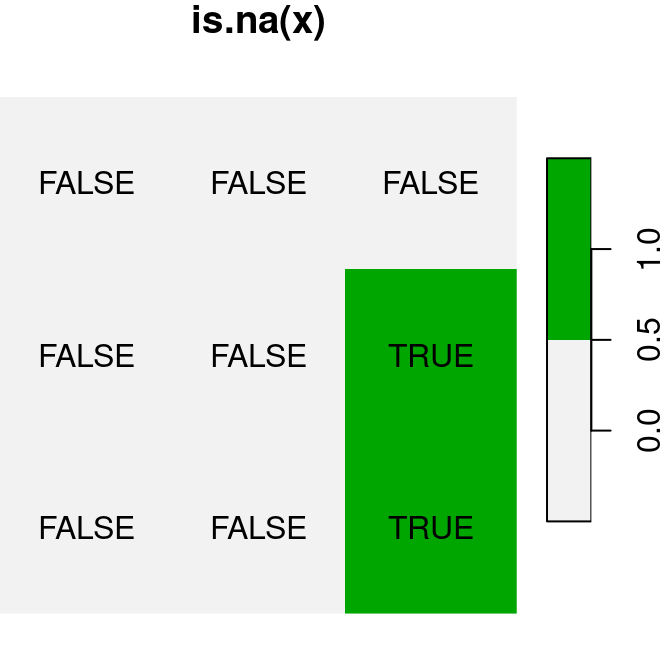
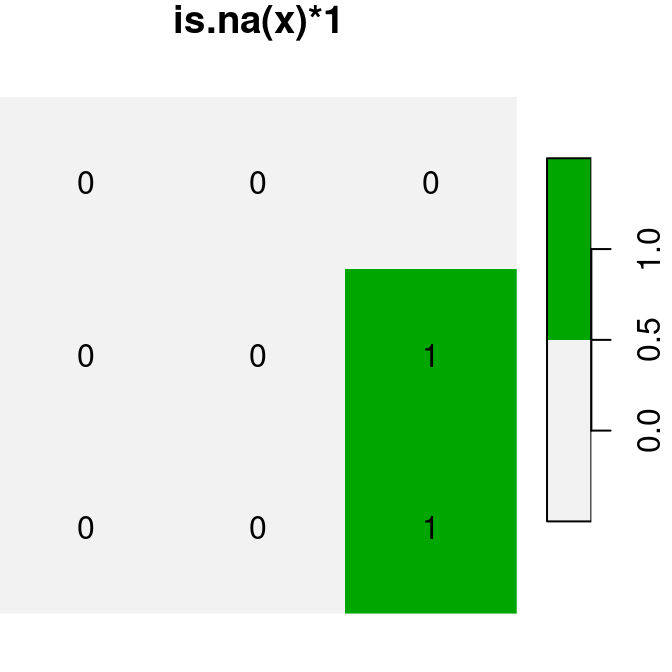
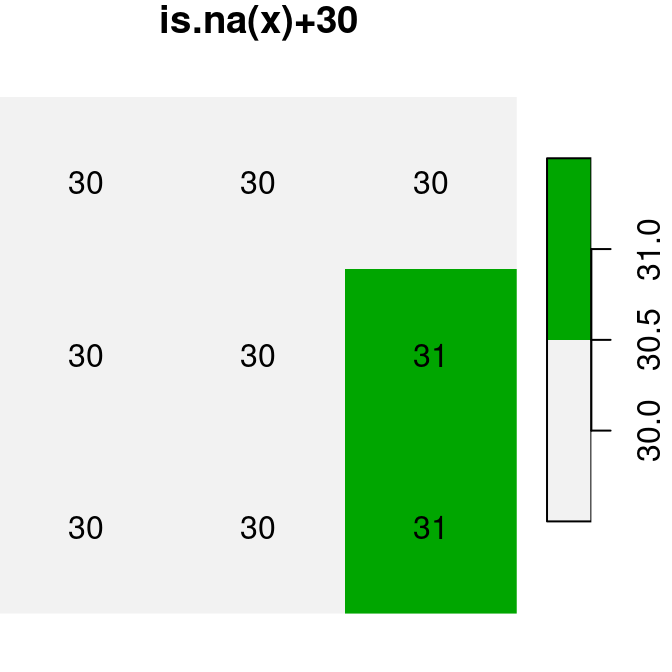
Figure 6.17: Conversion of raster values from logical to numeric
A logical raster can be used to assign new values into a subset of raster values. For example, we can use the logical raster is.na(x) to replace all NA values in the raster x with a new value:
The “before” and “after” images are shown in Figure 6.18:
plot(x, text_values = TRUE, breaks = seq(0.2, 0.3, 0.02), col = terrain.colors(5), main = 'Before')
plot(x1, text_values = TRUE, breaks = seq(0.2, 0.3, 0.02), col = terrain.colors(5), main = 'After')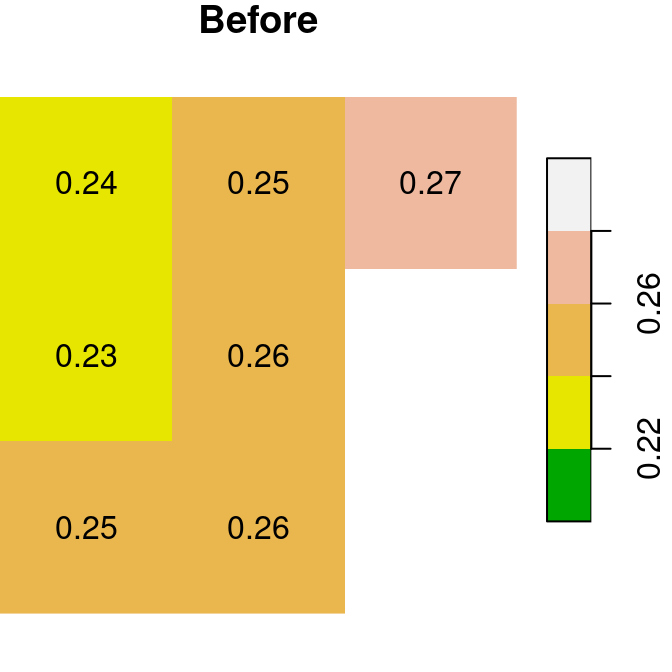
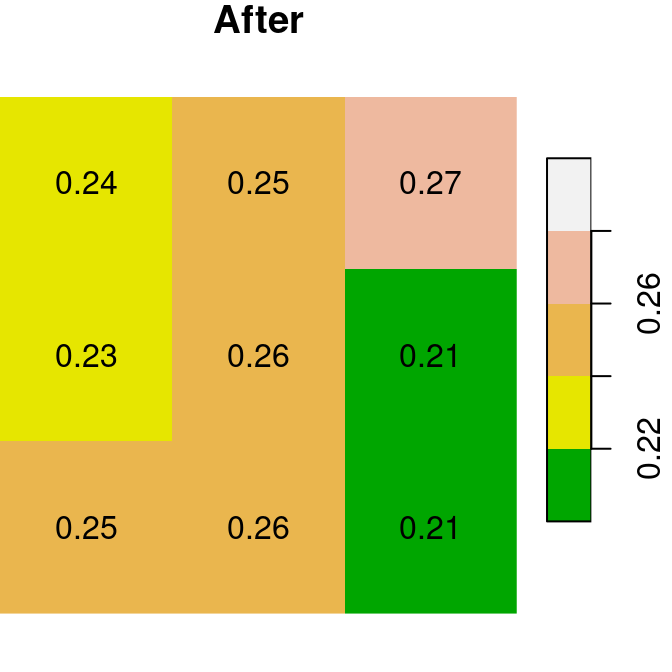
Figure 6.18: Assignmnent to raster subset using logical raster (before and after)
How can we replace the
NAvalues inxwith the average of all non-missing values?
How can we calculate the proportion of
NAvalues inx?
6.4.2 Landsat image
For the following example, let’s read another multi-band raster file named landsat_04_10_2000.tif. This is a part of a Landsat-5 satellite image with bands 1-5 and 7, after radiometric and atmospheric corrections. The raster values represent spectral reflectance, therefore all values are between 0 and 1.
We can assign meaningful layer names according to the spectral range that is captured by each band24:
bands = c('Blue', 'Green', 'Red', 'NIR', 'SWIR1', 'SWIR2')
l = st_set_dimensions(l, 'band', values = bands)
names(l) = 'reflectance'Protting the raster shows the reflectance images for each spectral band (Figure 6.19):
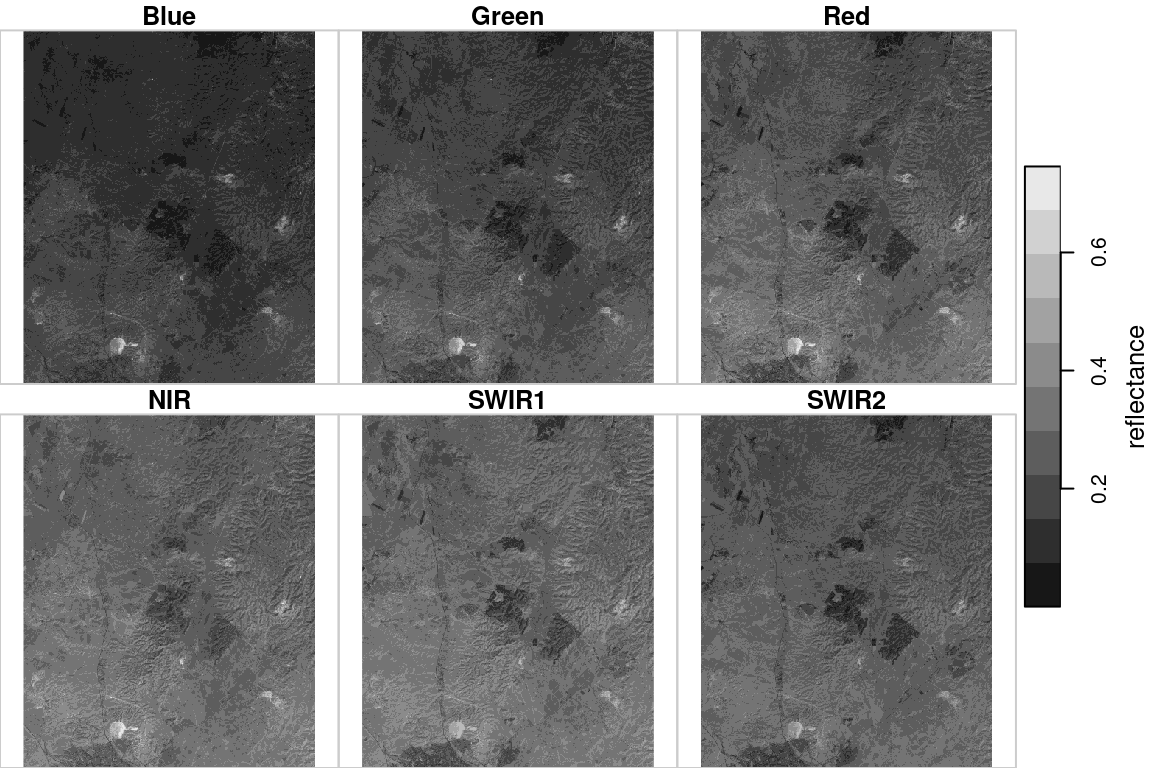
Figure 6.19: Landsat satellite image, bands 1-5 and 7
6.4.3 True color and false color images
An RGB image is an image where the colors are defines based on red, green, and blue intensity per pixel. An RGB image can therefore be constructed from red, green, and blue bands in a multispectral sensor (Figure 6.20), such as the ones in a digital camera or a satellite.
Figure 6.20: RGB image (https://datacarpentry.org/organization-geospatial/01-spatial-data-structures-formats/index.html)
A true color image is an image where the red, green and blue reflectance are red, green and blue colors. A true color image thus simulates the way that we would see the photographed area when looking from above, with the human eye. A false color image is an image where colors are mapped in any different way. A false color image can also include spectral bands that the human eye connot see, such as Near Infra Red (NIR).
True color and false color images can be produced with the plot function, specifying the red, green and blue bands with the rgb parameter. For example, in the Landsat image l, red, green and blue bands are 3, 2, and 1, therefore specifying rgb=c(3,2,1) produces a true color image (Figure 6.21). A different designation, rgb=c(4,3,2), produces a commonly used false color image where NIR is mapped to red, red is mapped to green and green is mapped to blue25 (Figure 6.21):

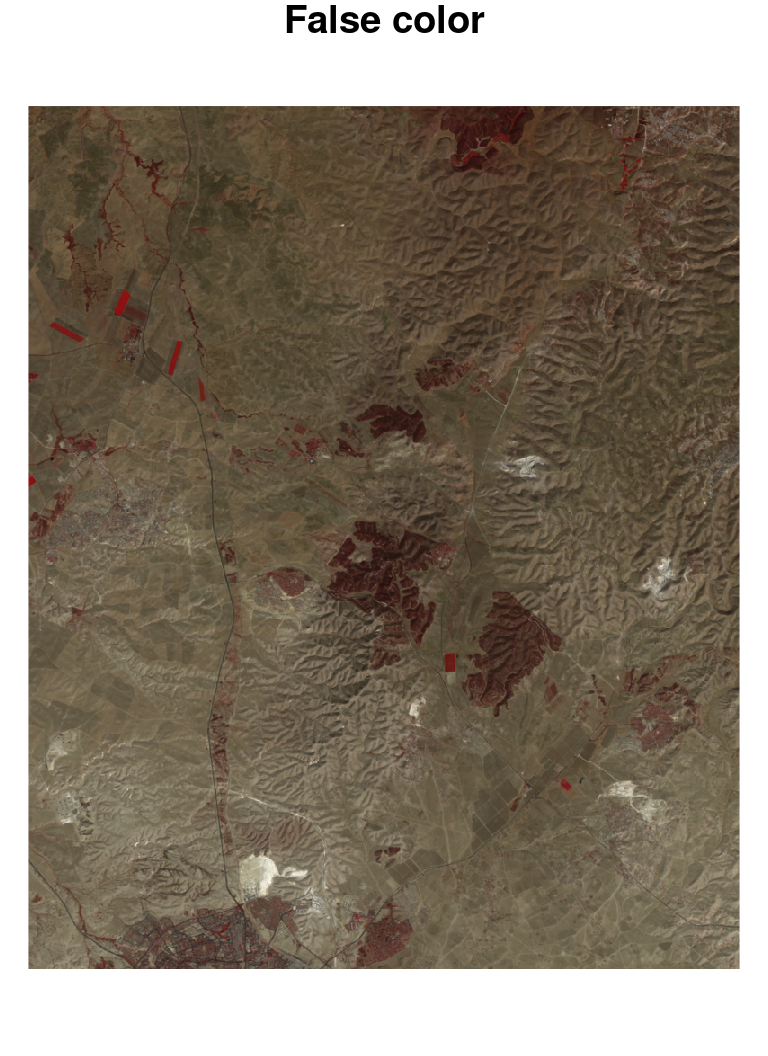
Figure 6.21: True color (left) and false color (right) images
6.4.4 Calculating NDVI
Multi-spectral images are commonly transformed to single band images using a spectral index. A common example of such an index is the NDVI, which we are familiar with from the raster MOD13A3_2000_2019.tif (Section 5.3.6). NDVI is defined as the difference between NIR and Red reflectance, divided by their sum (Equation (6.1)).
\[\begin{equation} NDVI=\frac{NIR-Red}{NIR + Red} \tag{6.1} \end{equation}\]
We can calculate an NDVI image based on the red and NIR bands in the Landsat image l, using raster algebra, as follows:
red = l[,,,3, drop = TRUE]
nir = l[,,,4, drop = TRUE]
ndvi = (nir - red) / (nir + red)
names(ndvi) = 'NDVI'The resulting NDVI image is shown in Figure 6.22:
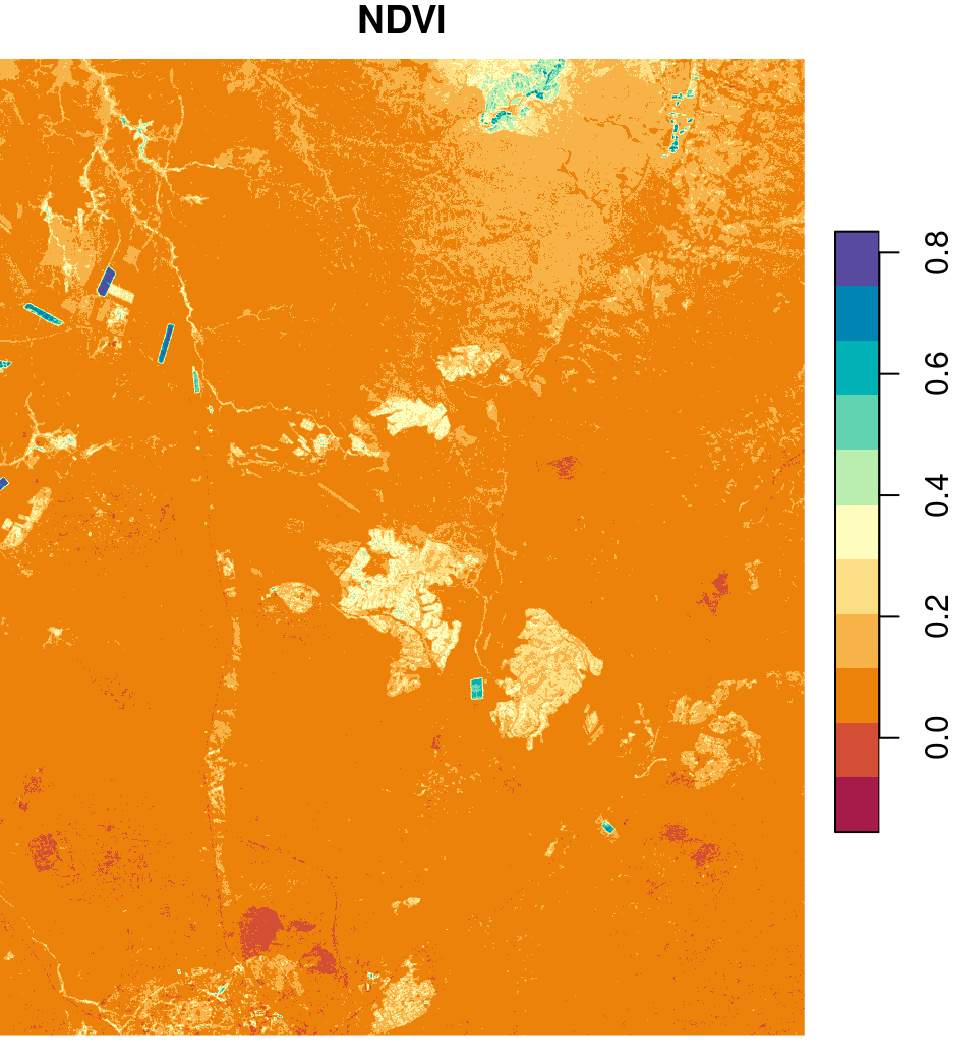
Figure 6.22: An NDVI image calculated based on a Landsat satellite image
6.5 Raster classification
Raster classification is the process of converting a continuous raster to a discrete one, by giving all pixels in a particular range of values a new uniform value (Figure 6.23).
Figure 6.23: Raster classification (http://rpubs.com/etiennebr/visualraster)
A raster can be classified, i.e., converted from a continuous raster to a categorical one, by assigning a new value to distinct ranges of the original values. For example, we can reclassify the continuous NDVI raster into two categories:
- Low (\(NDVI\leq0.2\)) get the value of
0 - High (\(NDVI>0.2\)) get the value of
1
Here is how we can do this with R code:
The result is shown in Figure 6.2426:
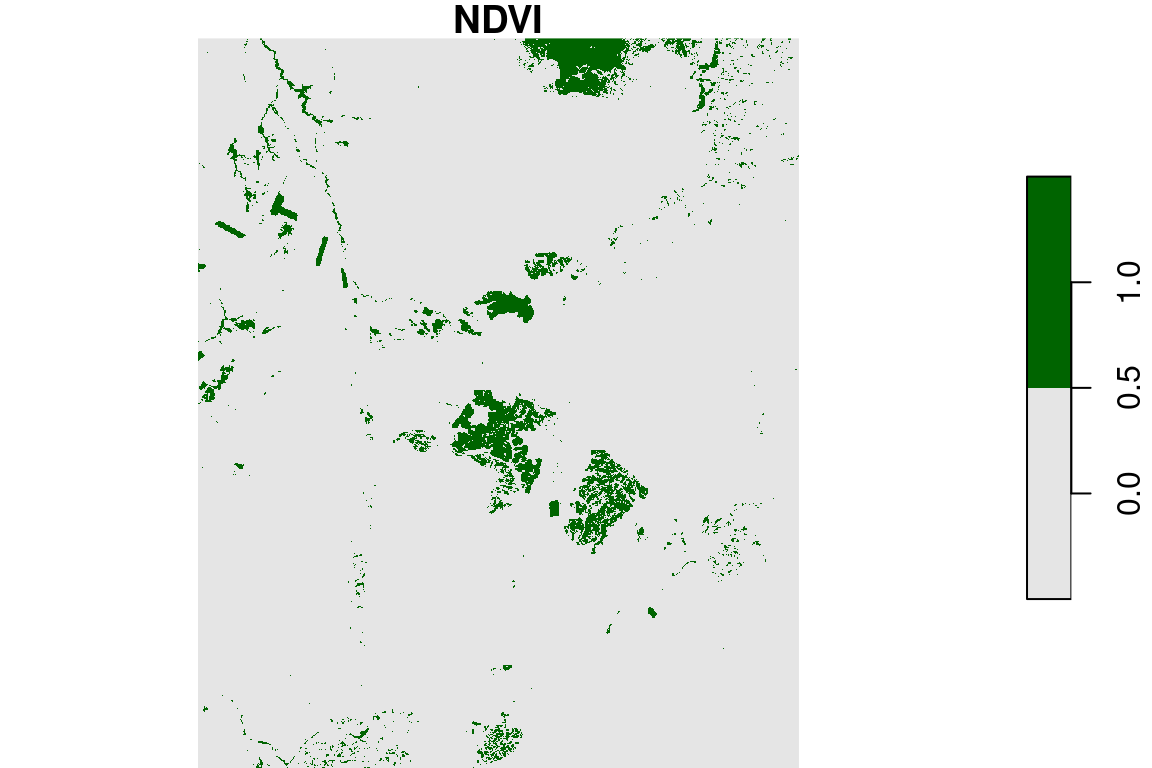
Figure 6.24: Reclassified NDVI image
6.6 Generalizing raster algebra with st_apply
6.6.1 Operating on each pixel
6.6.1.1 Introduction
Suppose that we have a small raster named s, with two layers:
s = r[, 99:101, 202:204, 1:2]
s[[1]]
## , , 1
##
## [,1] [,2] [,3]
## [1,] 0.2404 0.2313 0.2452
## [2,] 0.2513 0.2584 0.2564
## [3,] 0.2735 NA NA
##
## , , 2
##
## [,1] [,2] [,3]
## [1,] 0.2236 0.2375 0.2660
## [2,] 0.2680 0.2856 0.2563
## [3,] 0.2852 0.2746 0.2611and that we would like to add up the two layers, plus a constant value of 10. We have already seen this is a raster algebra operation that can be achieved with arithmetic operators (Section 6.4.1):
u = s[,,,1,drop=TRUE] + s[,,,2,drop=TRUE] + 10
u[[1]]
## [,1] [,2] [,3]
## [1,] 10.4640 10.4688 10.5112
## [2,] 10.5193 10.5440 10.5127
## [3,] 10.5587 NA NABut what if we want to apply a function on three, ten, or a hundred layers? Specifying each and every layer is impractical in such case. For that, we have a more general raster algebra approach using st_apply. The st_apply function is very similar to apply (Section 4.5). It takes an object, the dimension indices or names we wish to operate on, and a function, then applies the function on each subset along the dimension(s).
For example, the following expression calculates the same raster u as shown above, using st_apply:
u = st_apply(X = s, MARGIN = c('x', 'y'), FUN = function(x) sum(x) + 10)
u[[1]]
## [,1] [,2] [,3]
## [1,] 10.4640 10.4688 10.5112
## [2,] 10.5193 10.5440 10.5127
## [3,] 10.5587 NA NAwhere function(x) sum(x)+10 is a function applied on the vector of values x of each pixel.
The FUN parameter determines the function which is going to be applied on each subset of the chosen dimension, just like in apply (Section 4.5). In case the dimension we operate on is c('x','y'), i.e., “pixels” (Section 5.2.3), the FUN parameter determines the function which calculates each pixel value in the output raster, given the respective pixel values of the input raster from all layers. The FUN function needs to accept a vector of any length and return a vector of length 1. As a result, st_apply returns a single-band raster with the calculated values for each pixel27.
Note that we could write MARGIN=1:2 instead of MARGIN=c('x','y'). However, using dimension names instead of dimension indices is more meaningful (the intention of our code is more clear to the reader), and safer (our code does not depend on dimension order).
The st_apply function thus makes it possible to summarize raster dimension(s) properties using various built-in functions, such as mean, sum, min, max, etc., as well as custom functions (Section 3.3), such as function(x) sum(x)+10.
6.6.1.2 NDVI
As another example, the expression to calculate NDVI we have seen above (Section 6.4.4):
can be replaced with the following analogous expression using st_apply:
6.6.1.3 Pixel means
Calculating pixel means, across all raster layers, is a very common operation with raster time series. The following expression uses st_apply along with FUN=mean to calculate a new raster with the average NDVI values (during the period 2000-2019) per pixel, based on the raster r. The additional na.rm=TRUE argument is passed to the function (mean, in this case). This makes the calculation ignore NA values:
The resulting average NDVI raster is shown in Figure 6.25:
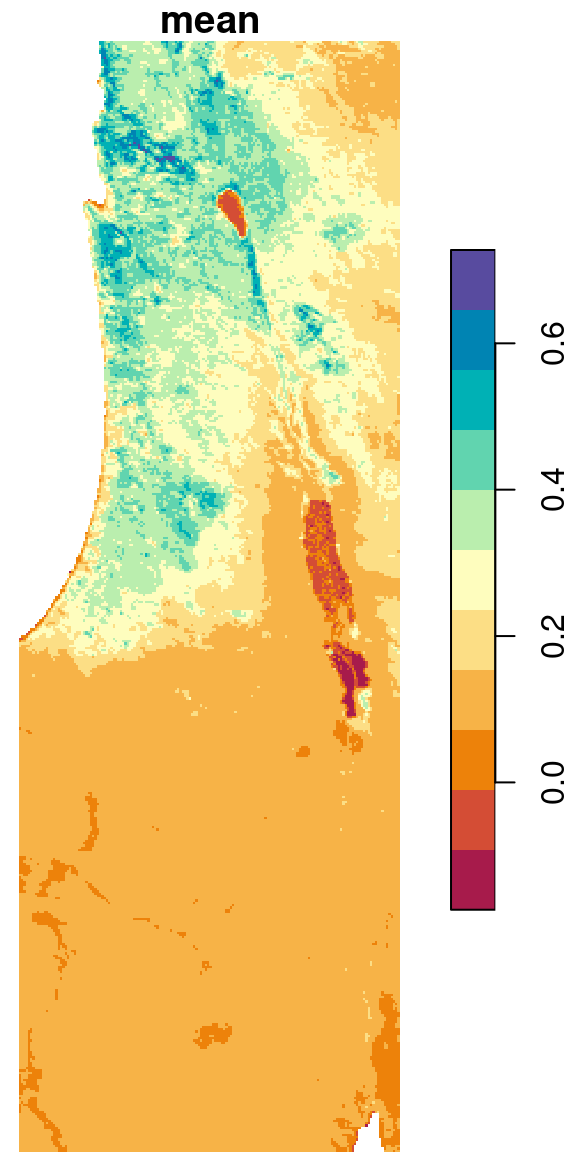
Figure 6.25: Average NDVI per pixel
6.6.1.4 Pixel minimum and maximum
In the following example, we use the min and max functions to calculate the minimal and maximal NDVI value, respectively, per pixel. In case our raster contains pixels where all values are NA, such as the case in r (6.6.1.6), we need to use slightly modified versions of min and max to avoid returning Inf and -Inf for those pixels:
The modified functions can be constructed as follows. In case all values are NA, the functions return NA. In case there is at least one non-NA value, the minimum or maximum is calculated:
min2 = function(x) if(all(is.na(x))) NA else min(x, na.rm = TRUE)
max2 = function(x) if(all(is.na(x))) NA else max(x, na.rm = TRUE)Let’s see if the new functions work as expected:
Now we can apply min2 and max2 on each pixel, to obtain single-band rasters with the minimum and maximum per pixel:
The results are shown in Figure 6.26:
plot(s_min, breaks = 'equal', col = hcl.colors(11, 'Spectral'), key.pos = 4)
plot(s_max, breaks = 'equal', col = hcl.colors(11, 'Spectral'), key.pos = 4)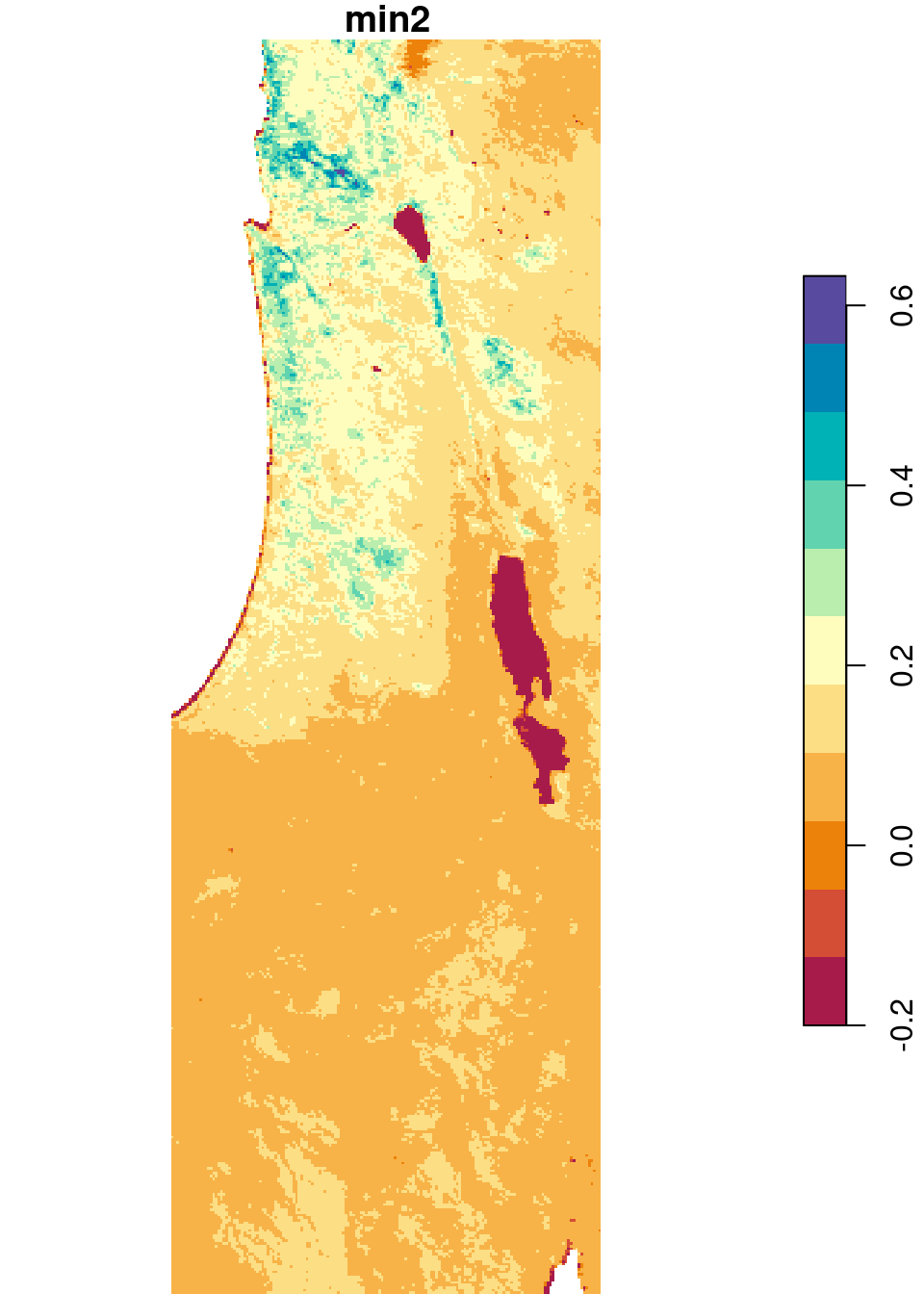
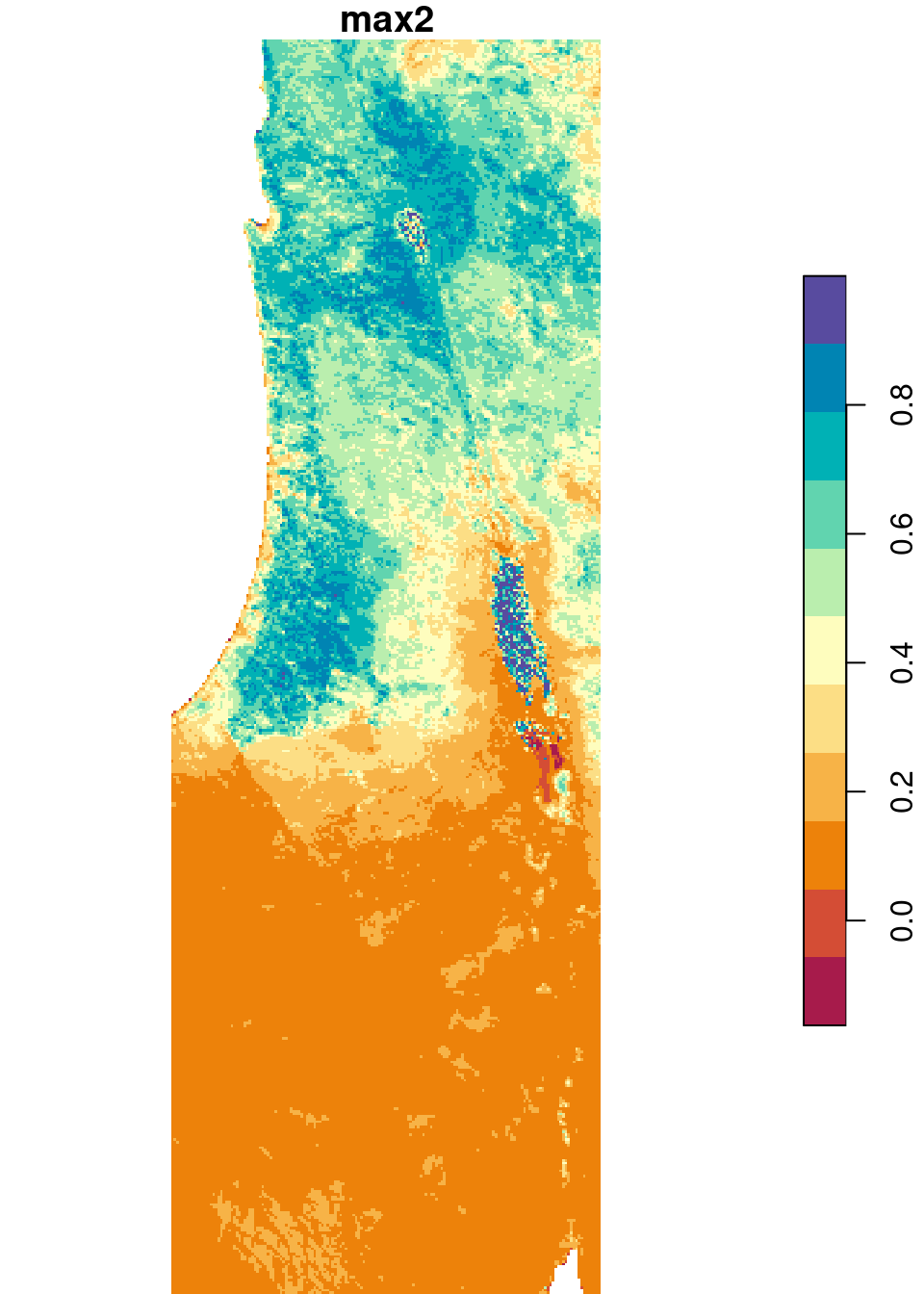
Figure 6.26: Minimum and maximum NDVI value per pixel
6.6.1.5 Pixel amplitudes
We can take the result from Section 6.6.1.4 and further calculate the difference between the maximum and minimum NDVI, i.e., the observed amplitude of NDVI values, in the raster r. This is yet another example of an arithmetic operator in raster algebra (Section 6.4.1):
The result is shown in Figure 6.27:
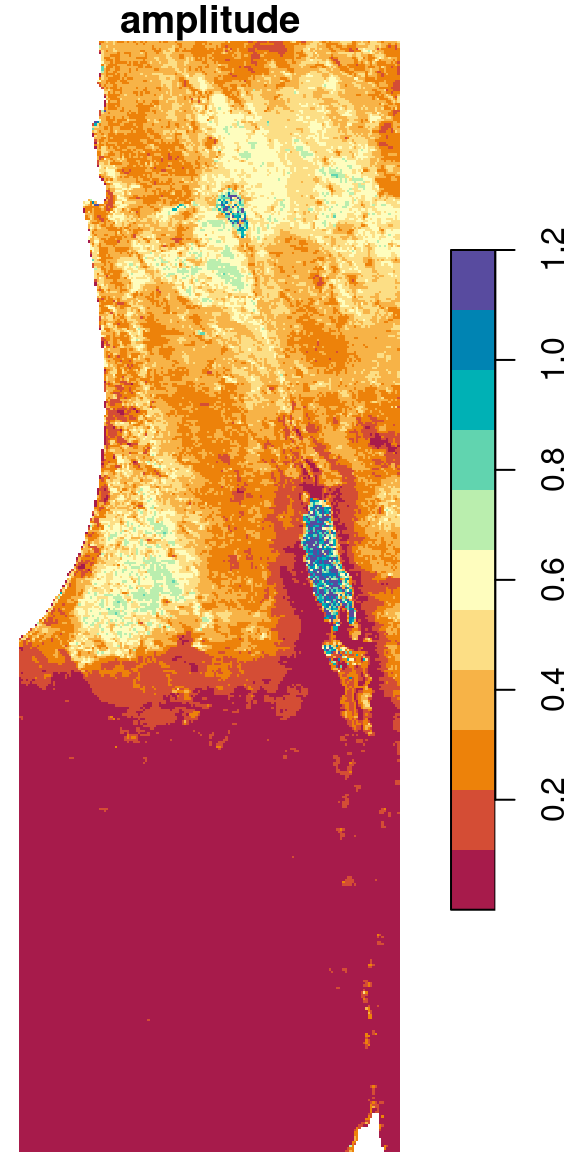
Figure 6.27: Amplitude of NDVI values per pixel
6.6.1.6 Pixel NA proportions
Another practical use case for st_apply is calculating the proportion of NA values per pixel:
The result is a raster with values between 0 and 1, where 0 means none of the 233 values of that pixel are NA, and 1 means that all 233 values are NA. The raster is shown in Figure 6.28. The second expression uses contour (Section 5.1.6) to highlight the areas where more than 5% of the pixel values are NA:
plot(s, breaks = 'equal', col = hcl.colors(100, 'Spectral'), reset = FALSE)
contour(s, levels = 0.05, add = TRUE)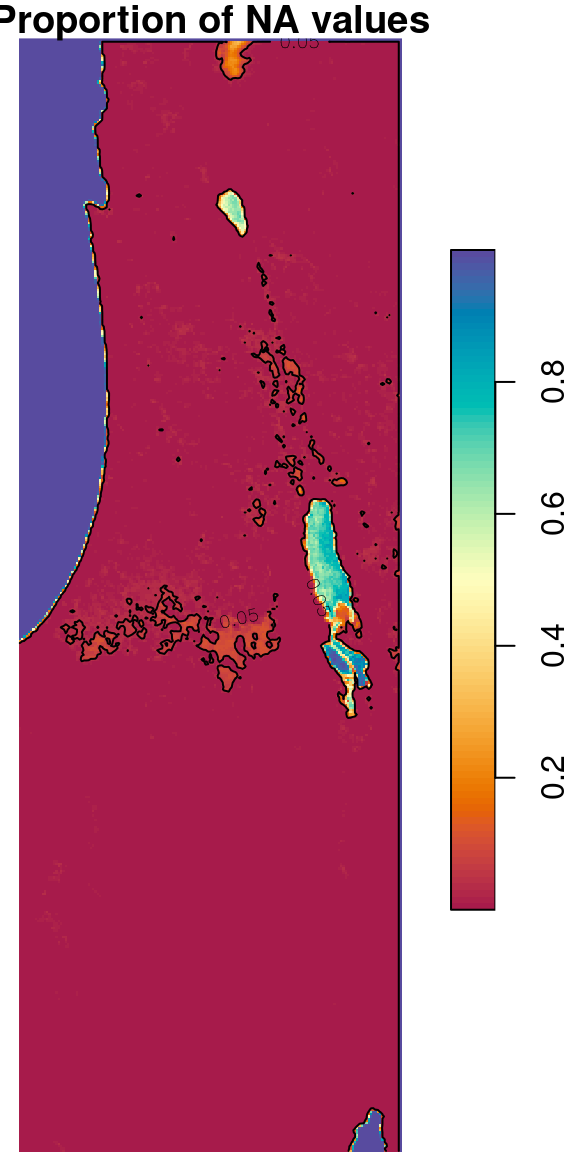
Figure 6.28: Proportion of NA values per pixel
6.6.2 Operating on each layer
In Section 6.6.1 we have seen examples of applying a function on the value of each pixel, using MARGIN=c('x','y'). Another useful mode of operation with st_apply is summarizing the properties of each layer, rather than each pixel. This is achieved with MARGIN='time'. For example, the following expressions calculate the minimum, mean and maximum NDVI value across the entire image per layer (i.e., per month):
layer_mean = st_apply(r, 'time', mean, na.rm = TRUE)
layer_min = st_apply(r, 'time', min, na.rm = TRUE)
layer_max = st_apply(r, 'time', max, na.rm = TRUE)The resulting stars objects are one-dimensional. Accordingly, layer_mean[[1]], layer_min[[1]] and layer_max[[1]] are one-dimensional arrays. One-dimensional arrays are treated as numeric vectors in most cases. For example, the following expression returns the average NDVI values of the first 10 layers in r:
layer_mean[[1]][1:10]
## [1] 0.2023428 0.2263248 0.2298939 0.1923323 0.1580042 0.1510594 0.1482469
## [8] 0.1443631 0.1496102 0.1650173The three resulting time series can therefore be visualized as follows (Figure 6.29):
times = st_get_dimension_values(r, 'time')
plot(times, layer_mean[[1]], type = 'o', ylim = range(r[[1]], na.rm = TRUE), xlab = 'Time', ylab = 'NDVI')
lines(times, layer_min[[1]], type = 'o', col = 'blue')
lines(times, layer_max[[1]], type = 'o', col = 'red')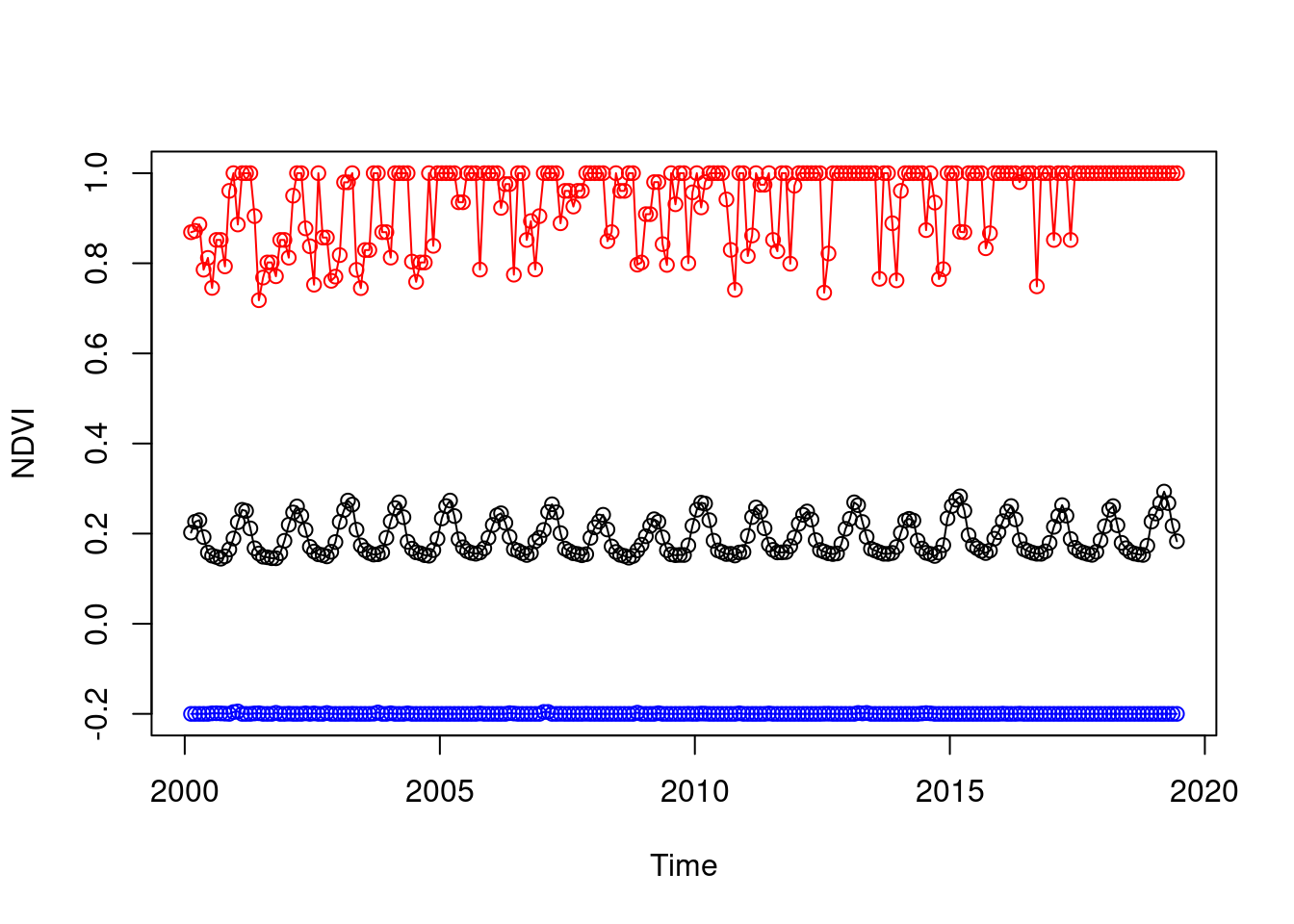
Figure 6.29: Minimum, mean and maximum NDVI over time
Note that
offsetanddeltashould be set together, since setting one of them resets the other toNA.↩︎https://landsat.usgs.gov/what-are-band-designations-landsat-satellites↩︎
This type of a false color image mapping emphasizes green vegetation (in red).↩︎
We can also do the above reclassification with a single expression, using
cut(ndvi, breaks = c(-Inf, 0.2, Inf)). This becomes especially convenient if we have numerous categories or classes. The result consists of values of typefactor, which we don’t go into in this book.↩︎st_applyalso accepts vector of (fixed) length n larger than 1, in which casest_applyreturns a multi-band raster with n layers.↩︎