Tables (pandas)#
Last updated: 2025-01-15 17:42:46
Introduction#
Tables are a fundamental data structure, common to many software interfaces, such as spreadsheet programs, relational databases, statistical software, etc. In general, a table is a two-dimensional structure where:
Columns represent variables
Rows represent observations
Accordingly, the data in a given table column is usually of the same type. For example, a text column may be used to store product names, and a numeric column may be used for storing product prices. Conversely, the data in a given row may comprise mixed types, such as the product name (text) and the price (a number) of a specific product.
In this chapter we introduce methods for working with tables in Python, through a popular third-party package named pandas, introducing two table-related data structures: Series and DataFrame. As we will see, the latter data structures are closely related to numpy, which was covered in the previous chapter (see Arrays (numpy)), since the data in each table column is actually stored as an “extended” one-dimensional numpy array. We are going to cover standard table-related operations, such as:
Creating tables (see Table from scratch and Reading from file)
Examining table properties (see DataFrame properties)
Modifying tables (see Renaming columns, Modifying the index, Sorting, Operators, and Creating new columns)
Subsetting tables (see Subsetting Series, Subsetting DataFrames, and DataFrame filtering)
Conversion to simpler data structures (see Series/DataFrame to ndarray and Conversion to list)
Plotting table data (see Plotting (pandas))
Exporting a table to file (see Writing DataFrame to file)
In the next chapter, we are going to discuss more advanced operations with tables, such as table aggregation and table joins (see Table reshaping and joins). Later on, we are going to learn about the geopandas package for working with vector layers, which is an extension of pandas (see Vector layers (geopandas) and Geometric operations).
What is pandas?#
pandas is a popular Python package which provides a flexible set of tools for working with tabular data. Since tables are fundamental in data analysis, pandas is one of the most important packages for data analysis in Python. Moreover, as we will see later on in the book (see Vector layers (geopandas)), a vector layer is represented by an “extended” table—a table that has a “geometry” column. Accordingly, the package for working with vector layers, named geopandas, is an extension of pandas.
The pandas package defines two fundamental data structures:
Series—A one-dimensional structure, representing a table columnDataFrame—A two-dimensional structure, represent a table
As we will see shortly, pandas is essentially an extension of numpy. A pandas table (a data structure named DataFrame) is a collection of Series representing the table columns, whereas Series are actually nothing more than labelled numpy arrays. To repeat, these two data structures therefore form a hierarchy: each column in a DataFrame is a Series.
Note
For those familiar with the R programming language, the term DataFrame will sound familiar, resembling R’s own data structure for representing tables (the data.frame). Indeed, pandas borrowed many ideas from the R language. A pairwise comparison of pandas vs. R functionality can be found in the pandas documentation Comparison with R / R libraries.
Table from scratch#
Creating a Series#
To understand how a DataFrame is structured, we will experiment with creating one from scratch. First we need to load the pandas package, as well as numpy which we use in some of the examples:
import numpy as np
import pandas as pd
A Series can be created, from a list, or from an ndarray, using pd.Series. For example, the following expression creates a Series from a list containing three strings, the names of three railway stations in southern Israel:
name = pd.Series(['Beer-Sheva Center', 'Beer-Sheva University', 'Dimona'])
name
0 Beer-Sheva Center
1 Beer-Sheva University
2 Dimona
dtype: object
Note
In pandas, strings are always stored as 'object' arrays (Series), unlike numpy which has specialized native data types for strings.
The following expression creates another Series, named lines, containing the number of railway lines going through the latter stations. This time, we create the Series from an ndarray:
lines = pd.Series(np.array([4, 5, 1]))
lines
0 4
1 5
2 1
dtype: int64
A Series object is essentially a numpy array, combined with indices. Here is how we can access each of these components, separately:
lines.to_numpy() ## Series values, as 'ndarray'
array([4, 5, 1])
lines.index ## Series index
RangeIndex(start=0, stop=3, step=1)
As you can see, Series values comprise a numpy array, which we are already familiar with. Series indices, however, comprise a special type of object used for indexing in pandas:
type(name.index)
pandas.core.indexes.range.RangeIndex
By default, the index is just a sequence of consecutive integers (such as in names and lines). When necessary, we can define any other sequence of values as a Series index (see Modifying the index), then use it to access specific values (see Subsetting Series).
Index objects are similar to arrays in many ways. For example, they have .shape and .dtype attributes, just like arrays do:
lines.index.shape
(3,)
lines.index.dtype
dtype('int64')
Creating a DataFrame#
While a Series can be thought of as an extended one-dimensional array, a DataFrame can be thought of as a collection of Series comprising table columns. Unlike a two-dimensional numpy array, a DataFrame can be composed of different types of values (among the columns). Consequently:
a
DataFramecolumn (i.e., aSeries) contains values of the same type, usually representing a particular variablea
DataFramerow may contain values of different types, usually representing a specific observation
Since a DataFrame is two-dimensional, it has two sets of idices, row and column indices, just like a two-dimensional array. It is important to note that all Series that comprise the columns of a DataFrame share the same index.
A DataFrame can be created from scratch, using the pd.DataFrame function. To do that, let us create four more Series which are going to comprise the DataFrame columns. Altogether, the DataFrame is going to represent various properties of three railway stations:
city = pd.Series(['Beer-Sheva', 'Beer-Sheva', 'Dimona'])
piano = pd.Series([False, True, False])
lon = pd.Series([34.798443, 34.812831, 35.011635])
lat = pd.Series([31.243288, 31.260284, 31.068616])
Here is a printout of the four Series we just created:
city
0 Beer-Sheva
1 Beer-Sheva
2 Dimona
dtype: object
piano
0 False
1 True
2 False
dtype: bool
lon
0 34.798443
1 34.812831
2 35.011635
dtype: float64
lat
0 31.243288
1 31.260284
2 31.068616
dtype: float64
Now, the six different Series can be combined into a DataFrame by passing them, as a dict (see Dictionaries (dict)), to the pd.DataFrame function. That way, the dict names will form the DataFrame column names, while the corresponding Series will form the column contents:
d = {
'name': name,
'city': city,
'lines': lines,
'piano': piano,
'lon': lon,
'lat': lat
}
stations = pd.DataFrame(d)
stations
| name | city | lines | piano | lon | lat | |
|---|---|---|---|---|---|---|
| 0 | Beer-Sheva Center | Beer-Sheva | 4 | False | 34.798443 | 31.243288 |
| 1 | Beer-Sheva University | Beer-Sheva | 5 | True | 34.812831 | 31.260284 |
| 2 | Dimona | Dimona | 1 | False | 35.011635 | 31.068616 |
Note
Note that there are other methods to create a DataFrame, passing different types of input to the pd.DataFrame function. For example, another useful method of creating a DataFrame by passing a two-dimensional array to pd.DataFrame.
DataFrame row indices are analogous to Series indices, and accessed exactly the same way:
stations.index
RangeIndex(start=0, stop=3, step=1)
Additionally, a DataFrame has column indices, which function as column names and can be accessed through the .columns property:
stations.columns
Index(['name', 'city', 'lines', 'piano', 'lon', 'lat'], dtype='object')
Reading from file#
In practice, we rarely need to create a DataFrame from scratch. More often, we read a table from a file with tabular data, such as a CSV file.
The pd.read_csv function is used to read a CSV file into a DataFrame in our Python environment. For example, the following expression reads the CSV file named 'ZonAnn.Ts+dSST.csv', which contains global temperature data from GISS. The values are anomalies, i.e., deviations from the 1951-1980 mean, in the years 1880-2023 ('Year'), both globally ('Glob') and for specific latitudes ('NHem'=Nothern Hemishpere, '24N-90N' latitudes 24N-90N, etc.):
dat = pd.read_csv('data/ZonAnn.Ts+dSST.csv')
dat
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 | -0.38 | ... | -0.14 | -0.11 | -0.04 | 0.05 | 0.67 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 | -0.36 | ... | 0.11 | 0.10 | -0.06 | -0.07 | 0.60 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 | -0.31 | ... | -0.04 | -0.05 | 0.01 | 0.04 | 0.63 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 | -0.34 | ... | -0.17 | -0.16 | -0.04 | 0.07 | 0.50 |
| 4 | 1884 | -0.28 | -0.42 | -0.15 | -0.61 | ... | -0.12 | -0.17 | -0.19 | -0.02 | 0.65 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 2019 | 0.98 | 1.20 | 0.75 | 1.42 | ... | 0.90 | 0.90 | 0.75 | 0.39 | 0.83 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 | 1.67 | ... | 0.88 | 0.84 | 0.58 | 0.39 | 0.89 |
| 141 | 2021 | 0.85 | 1.14 | 0.56 | 1.42 | ... | 0.72 | 0.60 | 0.72 | 0.32 | 0.30 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 | 1.52 | ... | 0.62 | 0.51 | 0.79 | 0.38 | 1.09 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 | 1.78 | ... | 1.07 | 1.04 | 0.91 | 0.44 | 0.63 |
144 rows × 15 columns
Note
pandas has several functions named pd.read_* to read formats other than CSV, such as pd.read_excel, pd.read_sql, pd.read_json, pd.read_spss, and pd.read_stata. Similarly, DataFrame objects have multiple methods (see Writing DataFrame to file) to write to various output formats. See the IO tools section in the pandas user guide for an overview of the input/output capabilities.
Note that rows and columns are labelled using indices, as shown above. In this case:
Rows are labelled with consecutive integers
Columns are labelled with column names, which are imported from the first line in CSV file
DataFrame properties#
Overview#
One of the first things we may want to do with a DataFrame imported from a file is to examine its properties, as shown in the next few sections:
DataFrame dimensions#
DataFrame dimensions are accessible through the .shape property, just like in a numpy array (see Array dimensions). For example, dat (with the temperature data) has 144 rows and 15 columns:
dat.shape
(144, 15)
DataFrame column names#
The DataFrame column names can be obtained through the .columns property. Recall that this is a special data structure representing pandas indices (see Table from scratch):
dat.columns
Index(['Year', 'Glob', 'NHem', 'SHem', '24N-90N', '24S-24N', '90S-24S',
'64N-90N', '44N-64N', '24N-44N', 'EQU-24N', '24S-EQU', '44S-24S',
'64S-44S', '90S-64S'],
dtype='object')
Exercise 05-a
From your experience with Python so far, can you guess how can the columns names be transformed to a plain
list? (You can also try searching for the answer online.)
DataFrame column types#
The data types (see Data types) of the columns are contained in the .dtypes property:
dat.dtypes
Year int64
Glob float64
NHem float64
SHem float64
24N-90N float64
...
EQU-24N float64
24S-EQU float64
44S-24S float64
64S-44S float64
90S-64S float64
Length: 15, dtype: object
Note that this is a Series (of dtype objects). This means we can get a specific value by index using the ordinary Series subsetting methods which we learn about later on (see Subsetting Series). For example, the 'Year' column was imported into a Series of type int64:
dat.dtypes.loc['Year']
dtype('int64')
while the 'Glob' column (global temperature anomaly) was imported into a float64 column:
dat.dtypes.loc['Glob']
dtype('float64')
The basic pandas data types are in agreement with numpy data types (see Data types). For example, the dat table contains int64 and float64 data types. However, pandas and its extensions (such as geopandas, see What is geopandas?) also extend the numpy functionality with new data types (such as GeometryDtype, see GeoSeries (geometry) column).
Renaming columns#
Sometimes it is necessary to rename DataFrame columns. For example, we may wish to use shorter names which are easier to type, or make the columns match another table we are working with. Columns can be renamed using the .rename method, which accepts a columns argument of the form {'old_name':'new_name',...}. For example, here is how we can replace the 'Year' column name with lowercase 'year':
dat.rename(columns={'Year': 'year'})
| year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 | -0.38 | ... | -0.14 | -0.11 | -0.04 | 0.05 | 0.67 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 | -0.36 | ... | 0.11 | 0.10 | -0.06 | -0.07 | 0.60 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 | -0.31 | ... | -0.04 | -0.05 | 0.01 | 0.04 | 0.63 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 | -0.34 | ... | -0.17 | -0.16 | -0.04 | 0.07 | 0.50 |
| 4 | 1884 | -0.28 | -0.42 | -0.15 | -0.61 | ... | -0.12 | -0.17 | -0.19 | -0.02 | 0.65 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 2019 | 0.98 | 1.20 | 0.75 | 1.42 | ... | 0.90 | 0.90 | 0.75 | 0.39 | 0.83 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 | 1.67 | ... | 0.88 | 0.84 | 0.58 | 0.39 | 0.89 |
| 141 | 2021 | 0.85 | 1.14 | 0.56 | 1.42 | ... | 0.72 | 0.60 | 0.72 | 0.32 | 0.30 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 | 1.52 | ... | 0.62 | 0.51 | 0.79 | 0.38 | 1.09 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 | 1.78 | ... | 1.07 | 1.04 | 0.91 | 0.44 | 0.63 |
144 rows × 15 columns
Note
Another column renaming technique is to assign a list of strings of the same length as the number of columns to the .columns attribute, as in dat.columns=['col1','col2',...].
Subsetting in pandas#
Overview#
There are numerous methods of subsetting Series and DataFrames in pandas. For example, Series values can be subsetted using the [ operator. However, this can be confusing since both using numeric indices and the index values are permitted (at the time of writing), and it can be unclear which method is actually employed.
In agreement with the Python philosophy, where explicit is better than implicit, the recommended subsetting methods are using .loc and .iloc, which are discussed in the following sub-sections. The principal difference between .loc and .iloc is:
In this book, we are going to use .iloc and .loc methods when subsetting Series (this section) and DataFrame (see Subsetting DataFrames) objects. Nevertheless, for shorter syntax, we are going to use the [ shortcut in the following cases:
Selecting
DataFramecolumns using the[operator and a string, or alistof strings, as in:dat['Year']instead ofdat.loc[:,'Year'](see Selecting DataFrame columns)dat[['Year']]instead ofdat.loc[:,['Year']](see Selecting DataFrame columns)
Filtering
DataFramerows using the[operator and a booleanSeries, as in:dat[dat['Year']>2017]instead ofdat.loc[dat['Year']>2017](see DataFrame filtering)
Subsetting Series#
As mentioned above, the recommended way to subset Series is using the specialized methods named .loc and .iloc:
.loc—For subsetting using theSeriesindex.iloc—For subsetting using the (implicit)numpyindex
What follows after the method are the indices, inside square brackets ([), whereas the indices can be one of:
An individual value, such as
.loc['a']or.iloc[0]A slice, such as
.loc['a':'b']or.iloc[1:2]A
list, such as.loc[['a','c']]or.iloc[[0,2]]
For the next few examples, let us create a Series object named s, with:
values
11,12,13,14, andindices
'a','b','c','d',
using the index parameter of pd.Series:
s = pd.Series([11, 12, 13, 14], index=['a', 'b', 'c', 'd'])
s
a 11
b 12
c 13
d 14
dtype: int64
Here is how we can use .loc to select values using the three above-mentioned types of indices:
s.loc['a'] ## Individual index
np.int64(11)
s.loc['b':'c'] ## Slice
b 12
c 13
dtype: int64
s.loc[['a', 'c']] ## 'list' of indices
a 11
c 13
dtype: int64
And here is how we can use .iloc to select values using numpy-style numeric indices:
s.iloc[0] ## Individual index
np.int64(11)
s.iloc[1:2] ## Slice
b 12
dtype: int64
s.iloc[[0, 2]] ## 'list' of indices
a 11
c 13
dtype: int64
In the above examples, note how using an individual index returns a standalone value, while using a slice, or a list of indices, return a Series, even if the list or the slice contains just one element:
s.iloc[[0]]
a 11
dtype: int64
Also note that a slice using indices (such as s.loc['a':'b']) is inclusive, while a slice using implicit indices (such as .iloc[0:1]) excludes the last index similarly to list and numpy behavior.
Subsetting DataFrames#
Selecting DataFrame columns#
A DataFrame is a two-dimensional object, unlike a Series which is one-dimensional. Accordingly, the .loc and iloc methods of a DataFrame accept two indices, separated by a comma:
The first index refers to rows
The second index refers to columns
When we want to select all rows or columns, we place : in the respective index, similarly to numpy array subsetting (see Subsetting arrays). Using an individual index returns a Series, while using a slice or a list of indices—even if length 1—returns a DataFrame.
For example, this is how we can use .loc to extract one DataFrame column as a Series:
dat.loc[:, 'Glob']
0 -0.17
1 -0.09
2 -0.11
3 -0.17
4 -0.28
...
139 0.98
140 1.01
141 0.85
142 0.89
143 1.17
Name: Glob, Length: 144, dtype: float64
and here is how we can extract a single column as a DataFrame:
dat.loc[:, ['Glob']]
| Glob | |
|---|---|
| 0 | -0.17 |
| 1 | -0.09 |
| 2 | -0.11 |
| 3 | -0.17 |
| 4 | -0.28 |
| ... | ... |
| 139 | 0.98 |
| 140 | 1.01 |
| 141 | 0.85 |
| 142 | 0.89 |
| 143 | 1.17 |
144 rows × 1 columns
As mentioned above (see Overview), we can also use the shortcut [ operator to subset columns(s):
dat['Glob'] ## Shortcut for `dat.loc[:,'Glob']`
0 -0.17
1 -0.09
2 -0.11
3 -0.17
4 -0.28
...
139 0.98
140 1.01
141 0.85
142 0.89
143 1.17
Name: Glob, Length: 144, dtype: float64
dat[['Glob']] ## Shortcut for `dat.loc[:,['Glob']]`
| Glob | |
|---|---|
| 0 | -0.17 |
| 1 | -0.09 |
| 2 | -0.11 |
| 3 | -0.17 |
| 4 | -0.28 |
| ... | ... |
| 139 | 0.98 |
| 140 | 1.01 |
| 141 | 0.85 |
| 142 | 0.89 |
| 143 | 1.17 |
144 rows × 1 columns
Note
Dot notation, as in dat.Glob can also be used to select individual columns of a DataFrame. However, this is not recommended, because it does not work (1) with non-string column names, and (2) with column names that are the same as DataFrame methods, such as .pop.
We can pass a list of column names to select more then one column. For example:
dat[['Year', 'Glob', 'NHem', 'SHem']]
| Year | Glob | NHem | SHem | |
|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 |
| 4 | 1884 | -0.28 | -0.42 | -0.15 |
| ... | ... | ... | ... | ... |
| 139 | 2019 | 0.98 | 1.20 | 0.75 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 |
| 141 | 2021 | 0.85 | 1.14 | 0.56 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 |
144 rows × 4 columns
To select all columns except for the specified ones, we can use the .drop method combined with axis=1:
dat.drop(['Year', 'Glob', 'NHem', 'SHem', '24N-90N', '24S-24N', '90S-24S'], axis=1)
| 64N-90N | 44N-64N | 24N-44N | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|
| 0 | -0.80 | -0.52 | -0.26 | -0.14 | -0.11 | -0.04 | 0.05 | 0.67 |
| 1 | -0.93 | -0.50 | -0.19 | 0.11 | 0.10 | -0.06 | -0.07 | 0.60 |
| 2 | -1.41 | -0.31 | -0.13 | -0.04 | -0.05 | 0.01 | 0.04 | 0.63 |
| 3 | -0.18 | -0.58 | -0.24 | -0.17 | -0.16 | -0.04 | 0.07 | 0.50 |
| 4 | -1.30 | -0.66 | -0.45 | -0.12 | -0.17 | -0.19 | -0.02 | 0.65 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 2.71 | 1.43 | 0.99 | 0.90 | 0.90 | 0.75 | 0.39 | 0.83 |
| 140 | 2.88 | 1.81 | 1.19 | 0.88 | 0.84 | 0.58 | 0.39 | 0.89 |
| 141 | 2.05 | 1.35 | 1.26 | 0.72 | 0.60 | 0.72 | 0.32 | 0.30 |
| 142 | 2.34 | 1.50 | 1.27 | 0.62 | 0.51 | 0.79 | 0.38 | 1.09 |
| 143 | 2.58 | 1.87 | 1.47 | 1.07 | 1.04 | 0.91 | 0.44 | 0.63 |
144 rows × 8 columns
Another useful technique is to use .loc combined with slices of column names. For example, we can select all columns between two specified ones:
dat.loc[:, 'Year':'SHem']
| Year | Glob | NHem | SHem | |
|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 |
| 4 | 1884 | -0.28 | -0.42 | -0.15 |
| ... | ... | ... | ... | ... |
| 139 | 2019 | 0.98 | 1.20 | 0.75 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 |
| 141 | 2021 | 0.85 | 1.14 | 0.56 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 |
144 rows × 4 columns
or all columns before a specified one:
dat.loc[:, :'SHem']
| Year | Glob | NHem | SHem | |
|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 |
| 4 | 1884 | -0.28 | -0.42 | -0.15 |
| ... | ... | ... | ... | ... |
| 139 | 2019 | 0.98 | 1.20 | 0.75 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 |
| 141 | 2021 | 0.85 | 1.14 | 0.56 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 |
144 rows × 4 columns
or all columns after a specified one:
dat.loc[:, 'SHem':]
| SHem | 24N-90N | 24S-24N | 90S-24S | 64N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -0.05 | -0.38 | -0.13 | -0.02 | -0.80 | ... | -0.14 | -0.11 | -0.04 | 0.05 | 0.67 |
| 1 | 0.00 | -0.36 | 0.10 | -0.07 | -0.93 | ... | 0.11 | 0.10 | -0.06 | -0.07 | 0.60 |
| 2 | -0.01 | -0.31 | -0.04 | 0.01 | -1.41 | ... | -0.04 | -0.05 | 0.01 | 0.04 | 0.63 |
| 3 | -0.07 | -0.34 | -0.17 | -0.01 | -0.18 | ... | -0.17 | -0.16 | -0.04 | 0.07 | 0.50 |
| 4 | -0.15 | -0.61 | -0.14 | -0.14 | -1.30 | ... | -0.12 | -0.17 | -0.19 | -0.02 | 0.65 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 0.75 | 1.42 | 0.90 | 0.64 | 2.71 | ... | 0.90 | 0.90 | 0.75 | 0.39 | 0.83 |
| 140 | 0.68 | 1.67 | 0.86 | 0.57 | 2.88 | ... | 0.88 | 0.84 | 0.58 | 0.39 | 0.89 |
| 141 | 0.56 | 1.42 | 0.66 | 0.53 | 2.05 | ... | 0.72 | 0.60 | 0.72 | 0.32 | 0.30 |
| 142 | 0.62 | 1.52 | 0.57 | 0.70 | 2.34 | ... | 0.62 | 0.51 | 0.79 | 0.38 | 1.09 |
| 143 | 0.85 | 1.78 | 1.05 | 0.72 | 2.58 | ... | 1.07 | 1.04 | 0.91 | 0.44 | 0.63 |
144 rows × 12 columns
Again, note that in the last examples, the subset is inclusive, i.e., includes both the start and end columns.
Exercise 05-b
How can we select all columns after the specified one excluding itself?
Selecting DataFrame rows#
Rows can be selected using .loc or .iloc, similarly to the way we use those methods to select columns. The difference is that we specify the first index, instead of the second. For example:
dat.iloc[[0], :] ## 1st row
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 | -0.38 | ... | -0.14 | -0.11 | -0.04 | 0.05 | 0.67 |
1 rows × 15 columns
dat.iloc[0:3, :] ## 1st row (inclusive) to 4th row (exclusive)
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 | -0.38 | ... | -0.14 | -0.11 | -0.04 | 0.05 | 0.67 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 | -0.36 | ... | 0.11 | 0.10 | -0.06 | -0.07 | 0.60 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 | -0.31 | ... | -0.04 | -0.05 | 0.01 | 0.04 | 0.63 |
3 rows × 15 columns
dat.iloc[[0, 2], :] ## 1st and 3rd rows
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 | -0.38 | ... | -0.14 | -0.11 | -0.04 | 0.05 | 0.67 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 | -0.31 | ... | -0.04 | -0.05 | 0.01 | 0.04 | 0.63 |
2 rows × 15 columns
Exercise 05-c
Create a subset of
datwith the 1st, 3rd, and 5th rows.
Selecting rows and columns#
Both DataFrame dimensions can be subsetted at once, by passing two indices to .loc or to .iloc. For example, here we select a subset of dat with the first four rows and the first four columns (i.e., the top-left “corner” of the table), using .iloc:
dat.iloc[0:4, 0:4]
| Year | Glob | NHem | SHem | |
|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 |
The same can be acheived using separate steps, for example using .loc to select columns, and then .iloc to select rows:
dat.loc[:, 'Year':'SHem'].iloc[0:4, :]
| Year | Glob | NHem | SHem | |
|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 |
Selecting DataFrame values#
Using the above-mentioned methods, we can access individual values from a DataFrame in several ways. The clearest syntax is probably splitting the operation into two parts: first selecting the column, then selecting the index within the column. For example, here is how we can get the first value in the 'Year' column of dat:
dat['Year'].iloc[0]
np.int64(1880)
Exercise 05-d
How can we get the last value in the
'Year'column ofdat, without explicitly typing its index (e.g.142)?
Series/DataFrame to ndarray#
A Series or a DataFrame is accessible as an ndarray, through the .to_numpy method. For example:
dat['Year'].iloc[0:3].to_numpy() ## 'Series' to array
array([1880, 1881, 1882])
dat.iloc[0:3, 1:3].to_numpy() ## 'DataFrame' to array
array([[-0.17, -0.28],
[-0.09, -0.18],
[-0.11, -0.21]])
Note that a Series is translated to a one-dimensional array, while a DataFrame is translated to a two-dimensional array.
Conversion to list#
Series to list#
In some cases we may prefer to use basic Python methods to work with data contained in a Series or in a DataFrame. For that purpose, the Series or the DataFrame can be converted to a list.
A Series can be converted to a list by directly applying the .to_list method, which is analogous to the .tolist method of ndarray (see ndarray to list). For example, let us take a small Series of length 5:
s = dat['Glob'].iloc[0:5]
s
0 -0.17
1 -0.09
2 -0.11
3 -0.17
4 -0.28
Name: Glob, dtype: float64
Here is how the Series can be converted to a list, using the .to_list method:
s.to_list()
[-0.17, -0.09, -0.11, -0.17, -0.28]
DataFrame to list#
A DataFrame can also be converted to a list, in which case the list will be nested, with each column represented by an internal list. Let us take a small DataFrame with five rows and three columns as an example:
dat1 = dat[['Year', 'NHem', 'SHem']].iloc[0:5, :]
dat1
| Year | NHem | SHem | |
|---|---|---|---|
| 0 | 1880 | -0.28 | -0.05 |
| 1 | 1881 | -0.18 | 0.00 |
| 2 | 1882 | -0.21 | -0.01 |
| 3 | 1883 | -0.28 | -0.07 |
| 4 | 1884 | -0.42 | -0.15 |
To convert the DataFrame to a list, we can use the .to_records method (possibly with index=False to omit the index), followed by .tolist. The result is a list to tuples, where each tuple represents a row:
dat1.to_records(index=False).tolist()
[(1880, -0.28, -0.05),
(1881, -0.18, 0.0),
(1882, -0.21, -0.01),
(1883, -0.28, -0.07),
(1884, -0.42, -0.15)]
Modifying the index#
Overivew#
In the above examples, the Series and DataFrame we created got the default, consecutive integer, index. As we will see, the index plays an important role in many operations in pandas. Therefore, often we would like to set a more meaningful, custom index. For example, it usually makes sense to set the index of a table representing a time series (such as dat) to the time points (such as 'Year' values). Many pandas operations then utilize the index. For example, when plotting the data, the x-axis will show the time labels (see Line plots (pandas)).
To set a new index, we:
Assign to the
.indexproperty of aSeries(see Setting Series index)Use the
.set_indexmethod of aDataFrame(see Setting DataFrame index)
To reset the index, i.e., get back to the default consecutive integer index, we use the .reset_index method (see Resetting the index).
We demonstrate these three techniques in the following sub-sections.
Setting Series index#
Let us go back to the name series from the beginning of the chapter (Creating a Series):
name
0 Beer-Sheva Center
1 Beer-Sheva University
2 Dimona
dtype: object
The following expression sets the index of the name series as ['a','b','c'], by assigning a list into the .index property:
name.index = ['a', 'b', 'c']
name
a Beer-Sheva Center
b Beer-Sheva University
c Dimona
dtype: object
Setting DataFrame index#
The (row) index of a DataFrame can also be “manually” set by assignment to .index. However, a more useful scenario is where we want one of the columns in the DataFrame to serve as the index, while also removing it from the DataFrame columns to avoid duplication. This can be done using .set_index.
For example, the following expression changes the index of stations to the station names:
stations = stations.set_index('name')
stations
| city | lines | piano | lon | lat | |
|---|---|---|---|---|---|
| name | |||||
| Beer-Sheva Center | Beer-Sheva | 4 | False | 34.798443 | 31.243288 |
| Beer-Sheva University | Beer-Sheva | 5 | True | 34.812831 | 31.260284 |
| Dimona | Dimona | 1 | False | 35.011635 | 31.068616 |
Now, name is no longer a column, but the index of stations.
Resetting the index#
To reset the index of a Series or a DataFrame, we use the .reset_index method. As a result, the index will be replaced with the default consecutive integer sequence. In case we want to remove the information in the index altoghether, we use the drop=True option. Otherwise, the index will be “transferred” into a new column.
For example, here is how we remove the ['a','b','c'] index, to get the original default index in the name Series. Note that the information in the index is lost and we get back to the original Series, due to the drop=True option:
name = name.reset_index(drop=True)
name
0 Beer-Sheva Center
1 Beer-Sheva University
2 Dimona
dtype: object
And here is how we reset the row index in the DataFrame named stations. This time, we do not want to lose the information, therefore the index “goes back” to the 'name' column:
stations = stations.reset_index()
stations
| name | city | lines | piano | lon | lat | |
|---|---|---|---|---|---|---|
| 0 | Beer-Sheva Center | Beer-Sheva | 4 | False | 34.798443 | 31.243288 |
| 1 | Beer-Sheva University | Beer-Sheva | 5 | True | 34.812831 | 31.260284 |
| 2 | Dimona | Dimona | 1 | False | 35.011635 | 31.068616 |
Sorting#
A DataFrame can be sorted using the .sort_values method. The first parameter (by) accepts a column name, or a list of column names, to sort by.
For example, the following expression sorts the rows of dat according to the global temperature anomaly (in the 'Glob' column), from lowest to highest:
dat.sort_values('Glob')
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 29 | 1909 | -0.48 | -0.49 | -0.48 | -0.53 | ... | -0.44 | -0.53 | -0.37 | -0.52 | -0.57 |
| 24 | 1904 | -0.47 | -0.48 | -0.46 | -0.51 | ... | -0.44 | -0.50 | -0.37 | -0.49 | -1.32 |
| 37 | 1917 | -0.46 | -0.57 | -0.36 | -0.50 | ... | -0.68 | -0.59 | -0.23 | -0.09 | 0.06 |
| 30 | 1910 | -0.44 | -0.44 | -0.43 | -0.42 | ... | -0.48 | -0.51 | -0.33 | -0.45 | 0.16 |
| 31 | 1911 | -0.44 | -0.42 | -0.47 | -0.42 | ... | -0.42 | -0.45 | -0.43 | -0.53 | 0.05 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 137 | 2017 | 0.92 | 1.17 | 0.67 | 1.38 | ... | 0.86 | 0.78 | 0.76 | 0.35 | 0.54 |
| 139 | 2019 | 0.98 | 1.20 | 0.75 | 1.42 | ... | 0.90 | 0.90 | 0.75 | 0.39 | 0.83 |
| 136 | 2016 | 1.01 | 1.31 | 0.73 | 1.55 | ... | 0.95 | 1.07 | 0.67 | 0.25 | 0.38 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 | 1.67 | ... | 0.88 | 0.84 | 0.58 | 0.39 | 0.89 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 | 1.78 | ... | 1.07 | 1.04 | 0.91 | 0.44 | 0.63 |
144 rows × 15 columns
The default is to sort in ascending order. If we want the opposite, i.e., a descending order, we need to specify ascending=False:
dat.sort_values('Glob', ascending=False)
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 143 | 2023 | 1.17 | 1.49 | 0.85 | 1.78 | ... | 1.07 | 1.04 | 0.91 | 0.44 | 0.63 |
| 136 | 2016 | 1.01 | 1.31 | 0.73 | 1.55 | ... | 0.95 | 1.07 | 0.67 | 0.25 | 0.38 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 | 1.67 | ... | 0.88 | 0.84 | 0.58 | 0.39 | 0.89 |
| 139 | 2019 | 0.98 | 1.20 | 0.75 | 1.42 | ... | 0.90 | 0.90 | 0.75 | 0.39 | 0.83 |
| 137 | 2017 | 0.92 | 1.17 | 0.67 | 1.38 | ... | 0.86 | 0.78 | 0.76 | 0.35 | 0.54 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 31 | 1911 | -0.44 | -0.42 | -0.47 | -0.42 | ... | -0.42 | -0.45 | -0.43 | -0.53 | 0.05 |
| 30 | 1910 | -0.44 | -0.44 | -0.43 | -0.42 | ... | -0.48 | -0.51 | -0.33 | -0.45 | 0.16 |
| 37 | 1917 | -0.46 | -0.57 | -0.36 | -0.50 | ... | -0.68 | -0.59 | -0.23 | -0.09 | 0.06 |
| 24 | 1904 | -0.47 | -0.48 | -0.46 | -0.51 | ... | -0.44 | -0.50 | -0.37 | -0.49 | -1.32 |
| 29 | 1909 | -0.48 | -0.49 | -0.48 | -0.53 | ... | -0.44 | -0.53 | -0.37 | -0.52 | -0.57 |
144 rows × 15 columns
Plotting (pandas)#
Histograms (pandas)#
pandas has several methods to visualize the information in a Series or a DataFrame, which are actually shortcuts to matplotlib functions (see The matplotlib package). These methods are useful for quick visual inspection of the data.
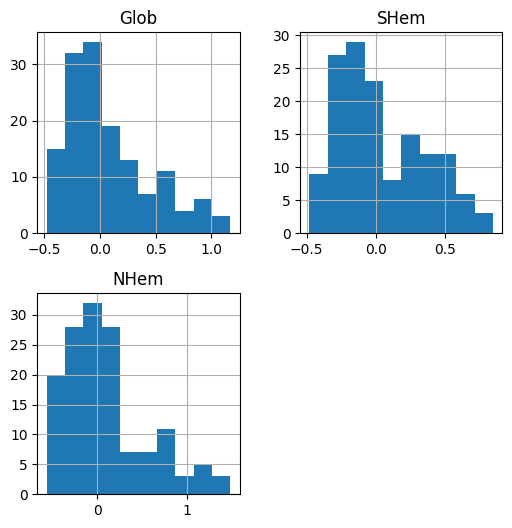
For example, we can draw a histogram of the column(s) in a DataFrame using the .hist method. For example:
dat[['Glob', 'SHem', 'NHem']].hist();
Line plots (pandas)#
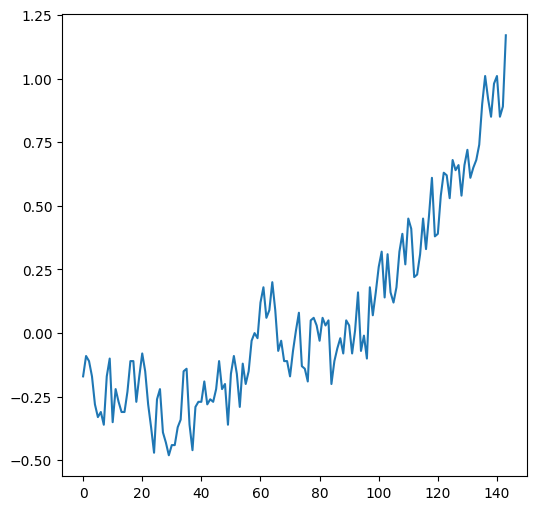
The .plot method, given a Series (or a DataFrame with one column), creates a line plot with the indices on the x-axis and the values on the y-axis:
dat['Glob'].plot();
In case we need to change the values on the x-axis, the simplest way is to set the required values as the index (see Modifying the index). For example, here we set the 'Year' column as the DataFrame index, then plot the 'Glob' column. As a result, the measurement year appears on the x-axis instead of the default consecutive index:
dat.set_index('Year')['Glob'].plot();
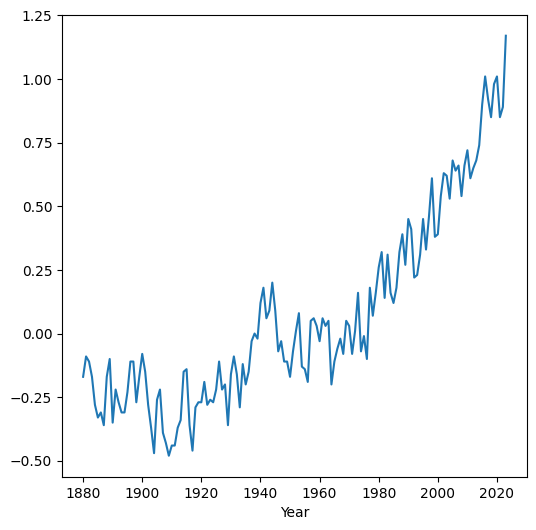
Pay attention to the order of operations (from left to right) in the above expression:
dat.set_index('Year')—Setting the index['Glob']—Extracting a column (asSeries).plot();—Plotting
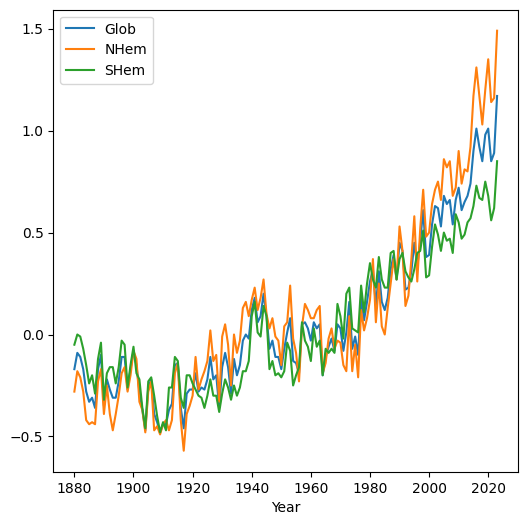
When we plot a DataFrame with more than one column, the columns are plotted as separate series in the same line plot. This is useful to compare the values in different columns. For example, plotting the 'Glob', 'NHem', and 'SHem' columns demonstrates that in recent years Northern Hemisphere ('NHem') temperatures have been relatively higher, while Southern Hemisphere ('SHem') temperatures have been relatively lower, compared to the global average ('Glob'):
dat.set_index('Year')[['Glob', 'NHem', 'SHem']].plot();
Scatterplots (pandas)#
One more useful type of plot is a scatterplot, where we display the association between two series in the form of scattered points. To produce a scatterplot, we use the .plot.scatter method of a DataFrame, specifying the names of the columns to be displayed in the x-axis (x) and y-axis (y).
For example, the following expression shows the relation between the values in the 'NHem' and 'SHem' columns of dat:
dat.plot.scatter(x='NHem', y='SHem');
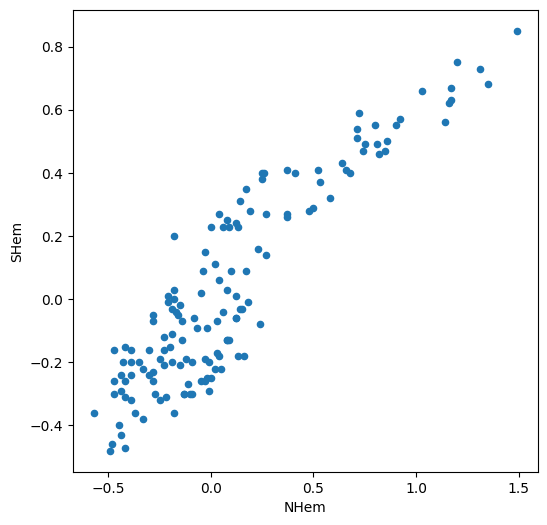
We can see a strong positive association between Northern and Southern Hemisphere temperatures across years. Namely, in years when the Nothern Hemisphere temperature is high, the Southern Hemisphere temperature also tends to be high, and vice versa.
Operators#
Operators on Series#
Operators and summary functions can be applied to Series objects, similarly to the way they can be applied to numpy arrays (see Vectorized operations and Summarizing array values, respectively). For example, summary methods such as .min, .max, and .mean can be applied on a Series to summarize its respective properties. Here is how we can find out the start and end 'Year' in dat:
dat['Year'].min()
np.int64(1880)
dat['Year'].max()
np.int64(2023)
and the average 'Glob' value:
dat['Glob'].mean()
np.float64(0.06770833333333333)
We can also combine Series with individual values, or combine two series, to apply pairwise arithmetic or boolean operators. For example, here is how we can subtract 1 from all 'Year' values:
dat['Year'] - 1
0 1879
1 1880
2 1881
3 1882
4 1883
...
139 2018
140 2019
141 2020
142 2021
143 2022
Name: Year, Length: 144, dtype: int64
and here is how we can calculate the yearly differences between the Northern Hemisphere and Southern Hemisphere temperature anomalies:
dat['NHem'] - dat['SHem']
0 -0.23
1 -0.18
2 -0.20
3 -0.21
4 -0.27
...
139 0.45
140 0.67
141 0.58
142 0.54
143 0.64
Length: 144, dtype: float64
Note that this is an arithmetic operation between two Series, which returns a new series.
Note
When performing operations between pairs of Series or DataFrames, pandas aligns the elements according to the index, rather than the position. When the data come from the same table (such as in the last example), the indices are guaranteed to match. However, when the data come from different tables, you must make sure their indices match.
Operators on DataFrames#
Operating on an entire DataFrame is more complex than operating on a Series (see Operators on Series). Accordingly, operators behave in different ways. For example, arithmetic and boolean operations combined with an individual value are applied per-element, resulting in a new DataFrame (assuming that all columns are numeric):
dat - 10
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1870 | -10.17 | -10.28 | -10.05 | -10.38 | ... | -10.14 | -10.11 | -10.04 | -9.95 | -9.33 |
| 1 | 1871 | -10.09 | -10.18 | -10.00 | -10.36 | ... | -9.89 | -9.90 | -10.06 | -10.07 | -9.40 |
| 2 | 1872 | -10.11 | -10.21 | -10.01 | -10.31 | ... | -10.04 | -10.05 | -9.99 | -9.96 | -9.37 |
| 3 | 1873 | -10.17 | -10.28 | -10.07 | -10.34 | ... | -10.17 | -10.16 | -10.04 | -9.93 | -9.50 |
| 4 | 1874 | -10.28 | -10.42 | -10.15 | -10.61 | ... | -10.12 | -10.17 | -10.19 | -10.02 | -9.35 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 2009 | -9.02 | -8.80 | -9.25 | -8.58 | ... | -9.10 | -9.10 | -9.25 | -9.61 | -9.17 |
| 140 | 2010 | -8.99 | -8.65 | -9.32 | -8.33 | ... | -9.12 | -9.16 | -9.42 | -9.61 | -9.11 |
| 141 | 2011 | -9.15 | -8.86 | -9.44 | -8.58 | ... | -9.28 | -9.40 | -9.28 | -9.68 | -9.70 |
| 142 | 2012 | -9.11 | -8.84 | -9.38 | -8.48 | ... | -9.38 | -9.49 | -9.21 | -9.62 | -8.91 |
| 143 | 2013 | -8.83 | -8.51 | -9.15 | -8.22 | ... | -8.93 | -8.96 | -9.09 | -9.56 | -9.37 |
144 rows × 15 columns
Methods such as .mean, however, are by default applied per column (i.e., axis=0):
dat.mean()
Year 1951.500000
Glob 0.067708
NHem 0.099444
SHem 0.036667
24N-90N 0.124722
...
EQU-24N 0.066181
24S-EQU 0.083056
44S-24S 0.047917
64S-44S -0.053681
90S-64S -0.074028
Length: 15, dtype: float64
We elaborate on this type of row- and column-wise operations later on (see Row/col-wise operations).
Creating new columns#
New DataFrame columns can be created by assignment of a Series to a non-existing column index. For example, the following expression calculates a Series of yearly differences between the Northern Hemisphere and Southern Hemisphere temperatures, as shown above, and assigns it to a new column named diff:
dat['diff'] = dat['NHem'] - dat['SHem']
dat
| Year | Glob | NHem | SHem | 24N-90N | ... | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | diff | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 | -0.38 | ... | -0.11 | -0.04 | 0.05 | 0.67 | -0.23 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 | -0.36 | ... | 0.10 | -0.06 | -0.07 | 0.60 | -0.18 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 | -0.31 | ... | -0.05 | 0.01 | 0.04 | 0.63 | -0.20 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 | -0.34 | ... | -0.16 | -0.04 | 0.07 | 0.50 | -0.21 |
| 4 | 1884 | -0.28 | -0.42 | -0.15 | -0.61 | ... | -0.17 | -0.19 | -0.02 | 0.65 | -0.27 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 2019 | 0.98 | 1.20 | 0.75 | 1.42 | ... | 0.90 | 0.75 | 0.39 | 0.83 | 0.45 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 | 1.67 | ... | 0.84 | 0.58 | 0.39 | 0.89 | 0.67 |
| 141 | 2021 | 0.85 | 1.14 | 0.56 | 1.42 | ... | 0.60 | 0.72 | 0.32 | 0.30 | 0.58 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 | 1.52 | ... | 0.51 | 0.79 | 0.38 | 1.09 | 0.54 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 | 1.78 | ... | 1.04 | 0.91 | 0.44 | 0.63 | 0.64 |
144 rows × 16 columns
Here is a plot (Line plots (pandas)) of the differences we just calulated, as function of time:
dat.set_index('Year')['diff'].plot();
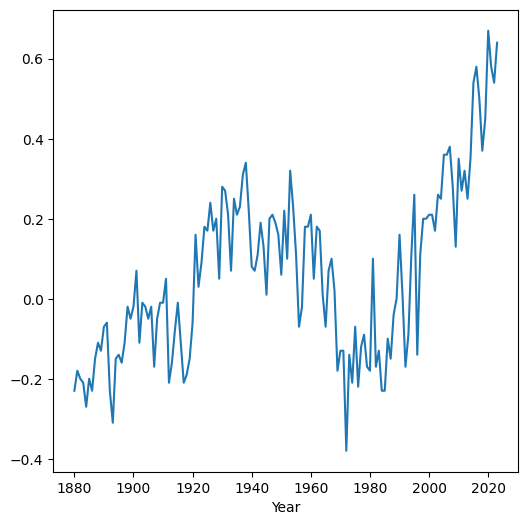
We can also assign an expression that combines series with individual values:
dat['NHem2'] = dat['NHem'] * 2
dat
| Year | Glob | NHem | SHem | 24N-90N | ... | 44S-24S | 64S-44S | 90S-64S | diff | NHem2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 | -0.38 | ... | -0.04 | 0.05 | 0.67 | -0.23 | -0.56 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 | -0.36 | ... | -0.06 | -0.07 | 0.60 | -0.18 | -0.36 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 | -0.31 | ... | 0.01 | 0.04 | 0.63 | -0.20 | -0.42 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 | -0.34 | ... | -0.04 | 0.07 | 0.50 | -0.21 | -0.56 |
| 4 | 1884 | -0.28 | -0.42 | -0.15 | -0.61 | ... | -0.19 | -0.02 | 0.65 | -0.27 | -0.84 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 2019 | 0.98 | 1.20 | 0.75 | 1.42 | ... | 0.75 | 0.39 | 0.83 | 0.45 | 2.40 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 | 1.67 | ... | 0.58 | 0.39 | 0.89 | 0.67 | 2.70 |
| 141 | 2021 | 0.85 | 1.14 | 0.56 | 1.42 | ... | 0.72 | 0.32 | 0.30 | 0.58 | 2.28 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 | 1.52 | ... | 0.79 | 0.38 | 1.09 | 0.54 | 2.32 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 | 1.78 | ... | 0.91 | 0.44 | 0.63 | 0.64 | 2.98 |
144 rows × 17 columns
or an individual value on its own (in which case it is duplicated across all rows):
dat['variable'] = 'temperature'
dat
| Year | Glob | NHem | SHem | 24N-90N | ... | 64S-44S | 90S-64S | diff | NHem2 | variable | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 | -0.38 | ... | 0.05 | 0.67 | -0.23 | -0.56 | temperature |
| 1 | 1881 | -0.09 | -0.18 | 0.00 | -0.36 | ... | -0.07 | 0.60 | -0.18 | -0.36 | temperature |
| 2 | 1882 | -0.11 | -0.21 | -0.01 | -0.31 | ... | 0.04 | 0.63 | -0.20 | -0.42 | temperature |
| 3 | 1883 | -0.17 | -0.28 | -0.07 | -0.34 | ... | 0.07 | 0.50 | -0.21 | -0.56 | temperature |
| 4 | 1884 | -0.28 | -0.42 | -0.15 | -0.61 | ... | -0.02 | 0.65 | -0.27 | -0.84 | temperature |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 2019 | 0.98 | 1.20 | 0.75 | 1.42 | ... | 0.39 | 0.83 | 0.45 | 2.40 | temperature |
| 140 | 2020 | 1.01 | 1.35 | 0.68 | 1.67 | ... | 0.39 | 0.89 | 0.67 | 2.70 | temperature |
| 141 | 2021 | 0.85 | 1.14 | 0.56 | 1.42 | ... | 0.32 | 0.30 | 0.58 | 2.28 | temperature |
| 142 | 2022 | 0.89 | 1.16 | 0.62 | 1.52 | ... | 0.38 | 1.09 | 0.54 | 2.32 | temperature |
| 143 | 2023 | 1.17 | 1.49 | 0.85 | 1.78 | ... | 0.44 | 0.63 | 0.64 | 2.98 | temperature |
144 rows × 18 columns
Let us delete the 'diff', 'NHem2', 'variable' columns we just created to get the original version of dat:
dat = dat.drop(['diff', 'NHem2', 'variable'], axis=1)
dat
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 | -0.38 | ... | -0.14 | -0.11 | -0.04 | 0.05 | 0.67 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 | -0.36 | ... | 0.11 | 0.10 | -0.06 | -0.07 | 0.60 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 | -0.31 | ... | -0.04 | -0.05 | 0.01 | 0.04 | 0.63 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 | -0.34 | ... | -0.17 | -0.16 | -0.04 | 0.07 | 0.50 |
| 4 | 1884 | -0.28 | -0.42 | -0.15 | -0.61 | ... | -0.12 | -0.17 | -0.19 | -0.02 | 0.65 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 2019 | 0.98 | 1.20 | 0.75 | 1.42 | ... | 0.90 | 0.90 | 0.75 | 0.39 | 0.83 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 | 1.67 | ... | 0.88 | 0.84 | 0.58 | 0.39 | 0.89 |
| 141 | 2021 | 0.85 | 1.14 | 0.56 | 1.42 | ... | 0.72 | 0.60 | 0.72 | 0.32 | 0.30 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 | 1.52 | ... | 0.62 | 0.51 | 0.79 | 0.38 | 1.09 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 | 1.78 | ... | 1.07 | 1.04 | 0.91 | 0.44 | 0.63 |
144 rows × 15 columns
DataFrame filtering#
Using conditional operators#
Most often, instead of selecting rows by index or by position (see Selecting DataFrame rows), we want to filter rows by a condition that we apply on table values. For example, suppose that we want to get a subset of dat with all rows after the year 2020. To do that, we can create a boolean Series, specifying which rows are to be retained:
sel = dat['Year'] > 2020
sel
0 False
1 False
2 False
3 False
4 False
...
139 False
140 False
141 True
142 True
143 True
Name: Year, Length: 144, dtype: bool
Then, the boolean series can be passed as an index inside square brackets ([), to select those rows corresponding to True:
dat[sel]
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 141 | 2021 | 0.85 | 1.14 | 0.56 | 1.42 | ... | 0.72 | 0.60 | 0.72 | 0.32 | 0.30 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 | 1.52 | ... | 0.62 | 0.51 | 0.79 | 0.38 | 1.09 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 | 1.78 | ... | 1.07 | 1.04 | 0.91 | 0.44 | 0.63 |
3 rows × 15 columns
The same can be acheived in a single step, without storing the Series in a variable such as sel, as follows:
dat[dat['Year'] > 2020]
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 141 | 2021 | 0.85 | 1.14 | 0.56 | 1.42 | ... | 0.72 | 0.60 | 0.72 | 0.32 | 0.30 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 | 1.52 | ... | 0.62 | 0.51 | 0.79 | 0.38 | 1.09 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 | 1.78 | ... | 1.07 | 1.04 | 0.91 | 0.44 | 0.63 |
3 rows × 15 columns
Here is an example with a more complex condition (note the parantheses!):
dat[(dat['Year'] > 2020) & (dat['Year'] <= 2022)]
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 141 | 2021 | 0.85 | 1.14 | 0.56 | 1.42 | ... | 0.72 | 0.60 | 0.72 | 0.32 | 0.30 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 | 1.52 | ... | 0.62 | 0.51 | 0.79 | 0.38 | 1.09 |
2 rows × 15 columns
To get the values of partucular column(s), we can use .loc, as follows:
dat.loc[(dat['Year'] > 2020) & (dat['Year'] <= 2022), 'Glob']
141 0.85
142 0.89
Name: Glob, dtype: float64
Exercise 05-e
Create a subset of
datwith all rows where the global temperature anomaly ('Glob') was above1degree.
Using .isin#
Sometimes, the condition we are interested in is not a comparison against an individual value (such as >2017, see Using conditional operators), but multiple values. For example, suppose we want to extract the rows corresponding to the years 2008, 2012, and 2016, in dat. One way is to compose a combined condition using |, using the numpy syntax we are familiar with (see numpy conditional operators). Again, recall that we must enclose each internal expression in breackets ( so that it is evaluated before |:
sel = (dat['Year'] == 2008) | (dat['Year'] == 2012) | (dat['Year'] == 2016)
dat[sel]
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 128 | 2008 | 0.54 | 0.68 | 0.40 | 0.91 | ... | 0.34 | 0.41 | 0.52 | 0.08 | 0.60 |
| 132 | 2012 | 0.65 | 0.81 | 0.49 | 1.04 | ... | 0.48 | 0.57 | 0.56 | 0.19 | 0.45 |
| 136 | 2016 | 1.01 | 1.31 | 0.73 | 1.55 | ... | 0.95 | 1.07 | 0.67 | 0.25 | 0.38 |
3 rows × 15 columns
As you can imagine, this method quickly becomes unfeasible if the number of items becomes large. Instead, we can use the .isin method, combined with a list of items to compare with the Series values:
sel = dat['Year'].isin([2008, 2012, 2016])
dat[sel]
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 128 | 2008 | 0.54 | 0.68 | 0.40 | 0.91 | ... | 0.34 | 0.41 | 0.52 | 0.08 | 0.60 |
| 132 | 2012 | 0.65 | 0.81 | 0.49 | 1.04 | ... | 0.48 | 0.57 | 0.56 | 0.19 | 0.45 |
| 136 | 2016 | 1.01 | 1.31 | 0.73 | 1.55 | ... | 0.95 | 1.07 | 0.67 | 0.25 | 0.38 |
3 rows × 15 columns
Recall the in operator which we learned about earlier (see The in operator). The in operator is analogous to the .isin method, but intended for lists.
Exercise 05-f
What do you think is the meaning of
dat[~sel]? Try it to check your answer.
Assignment to subsets#
When assigning to subsets of a column, we need to use the .loc method. Typically we combine:
A boolean
Seriesin the rows index, to select the observations we want to modifyAn
strin the columns index, to select the variable we want to modify
For example, the following expression sets the 'NHem' column value to 9999 in all years after 2020:
dat.loc[dat['Year'] > 2020, 'NHem'] = 9999
dat
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 | -0.38 | ... | -0.14 | -0.11 | -0.04 | 0.05 | 0.67 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 | -0.36 | ... | 0.11 | 0.10 | -0.06 | -0.07 | 0.60 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 | -0.31 | ... | -0.04 | -0.05 | 0.01 | 0.04 | 0.63 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 | -0.34 | ... | -0.17 | -0.16 | -0.04 | 0.07 | 0.50 |
| 4 | 1884 | -0.28 | -0.42 | -0.15 | -0.61 | ... | -0.12 | -0.17 | -0.19 | -0.02 | 0.65 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 2019 | 0.98 | 1.20 | 0.75 | 1.42 | ... | 0.90 | 0.90 | 0.75 | 0.39 | 0.83 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 | 1.67 | ... | 0.88 | 0.84 | 0.58 | 0.39 | 0.89 |
| 141 | 2021 | 0.85 | 9999.00 | 0.56 | 1.42 | ... | 0.72 | 0.60 | 0.72 | 0.32 | 0.30 |
| 142 | 2022 | 0.89 | 9999.00 | 0.62 | 1.52 | ... | 0.62 | 0.51 | 0.79 | 0.38 | 1.09 |
| 143 | 2023 | 1.17 | 9999.00 | 0.85 | 1.78 | ... | 1.07 | 1.04 | 0.91 | 0.44 | 0.63 |
144 rows × 15 columns
To get back to the original values, let us read the CSV file once again:
dat = pd.read_csv('data/ZonAnn.Ts+dSST.csv')
dat
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 | -0.38 | ... | -0.14 | -0.11 | -0.04 | 0.05 | 0.67 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 | -0.36 | ... | 0.11 | 0.10 | -0.06 | -0.07 | 0.60 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 | -0.31 | ... | -0.04 | -0.05 | 0.01 | 0.04 | 0.63 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 | -0.34 | ... | -0.17 | -0.16 | -0.04 | 0.07 | 0.50 |
| 4 | 1884 | -0.28 | -0.42 | -0.15 | -0.61 | ... | -0.12 | -0.17 | -0.19 | -0.02 | 0.65 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 2019 | 0.98 | 1.20 | 0.75 | 1.42 | ... | 0.90 | 0.90 | 0.75 | 0.39 | 0.83 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 | 1.67 | ... | 0.88 | 0.84 | 0.58 | 0.39 | 0.89 |
| 141 | 2021 | 0.85 | 1.14 | 0.56 | 1.42 | ... | 0.72 | 0.60 | 0.72 | 0.32 | 0.30 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 | 1.52 | ... | 0.62 | 0.51 | 0.79 | 0.38 | 1.09 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 | 1.78 | ... | 1.07 | 1.04 | 0.91 | 0.44 | 0.63 |
144 rows × 15 columns
Working with missing data (pandas)#
Missing data in pandas#
Missing values, denoting that the true value is unknown, are an inevitable feature of real-world data. In plain-text formats, such as CSV, missing values are commonly just “missing”, i.e., left blank, or marked with a text label such as 'NA'.
Treatment of “No Data” values in Series and DataFrames is derived from the underlying behavior in numpy (Working with missing data (numpy)), as pandas is based on numpy. Namely, “No Data” values are typically represented using np.nan.
pandas may automatically transform the Series data type, and “No Data” representation, for optimized data storage. For example, a Series of type int which contains one or more np.nan value, is automatically transformed to float, similarly to the behaviour we have seen with numpy arrays (Working with missing data (numpy)):
pd.Series([7, 4, 3])
0 7
1 4
2 3
dtype: int64
pd.Series([7, np.nan, 3])
0 7.0
1 NaN
2 3.0
dtype: float64
Moreover, None values are automatically transformed to np.nan (unlike numpy, where None values are preserved, resulting in an object array):
pd.Series([7, None, 3])
0 7.0
1 NaN
2 3.0
dtype: float64
“No Data” values are automatically assigned where necessary when creating a Series or a DataFrame through other methods too. For example, empty cells in a CSV file are assigned with np.nan when importing the CSV into a DataFrame (see Reading from file).
The 'education.csv' file contains the locations, and other details, about education facilities in Beer-Sheva:
'Institutio'—Institution name'type'—Institution type'ADDRESS'—Institution address'oldName'—Institution old name'lat'—Latitude'lon'—Longitude
If you open the CSV file in a spreadsheet program (such as Excel), you will see that some of the cells are empty. Those cells are assigned with np.nan when importing the table:
edu = pd.read_csv('data/education.csv')
edu
| Institutio | type | ADDRESS | oldName | lat | lon | |
|---|---|---|---|---|---|---|
| 0 | דוגית | גן ילדים | משה סתוי נ.זאב | NaN | 31.240226 | 34.767153 |
| 1 | שומרון | גן ילדים | שומרון שכ לון | NaN | 31.246547 | 34.765438 |
| 2 | שרת | גן ילדים | משה שרת 13 | NaN | 31.256681 | 34.766891 |
| 3 | חופית | גן ילדים | ניב דוד נ.זאב | NaN | 31.238660 | 34.770356 |
| 4 | רביבים | גן ילדים | מבצע משה נ.זאב | NaN | 31.237117 | 34.779331 |
| ... | ... | ... | ... | ... | ... | ... |
| 484 | ברעם | גן ילדים | נחל משמר | NaN | 31.233890 | 34.818100 |
| 485 | שילה | גן ילדים | נחל יהל 3 | NaN | 31.232486 | 34.821510 |
| 486 | קדם | גן ילדים | נחל יהל 3 | NaN | 31.232557 | 34.821390 |
| 487 | שמים | גן ילדים | וינגיט שכ ג | מיתרים | 31.254752 | 34.803860 |
| 488 | תכלת | גן ילדים | וינגיט שכ ג | מיתרים | 31.254470 | 34.804140 |
489 rows × 6 columns
First, let’s make the column names more convenient:
edu = edu.rename(columns={
'Institutio': 'name',
'ADDRESS': 'address',
'oldName': 'old_name'
})
edu
| name | type | address | old_name | lat | lon | |
|---|---|---|---|---|---|---|
| 0 | דוגית | גן ילדים | משה סתוי נ.זאב | NaN | 31.240226 | 34.767153 |
| 1 | שומרון | גן ילדים | שומרון שכ לון | NaN | 31.246547 | 34.765438 |
| 2 | שרת | גן ילדים | משה שרת 13 | NaN | 31.256681 | 34.766891 |
| 3 | חופית | גן ילדים | ניב דוד נ.זאב | NaN | 31.238660 | 34.770356 |
| 4 | רביבים | גן ילדים | מבצע משה נ.זאב | NaN | 31.237117 | 34.779331 |
| ... | ... | ... | ... | ... | ... | ... |
| 484 | ברעם | גן ילדים | נחל משמר | NaN | 31.233890 | 34.818100 |
| 485 | שילה | גן ילדים | נחל יהל 3 | NaN | 31.232486 | 34.821510 |
| 486 | קדם | גן ילדים | נחל יהל 3 | NaN | 31.232557 | 34.821390 |
| 487 | שמים | גן ילדים | וינגיט שכ ג | מיתרים | 31.254752 | 34.803860 |
| 488 | תכלת | גן ילדים | וינגיט שכ ג | מיתרים | 31.254470 | 34.804140 |
489 rows × 6 columns
For example, the old name of the first entry is unknown:
edu['old_name'].iloc[0]
nan
Detecting with pd.isna#
To detect “No Data” values, the .isna method can be applied on a Series or a DataFrame. The result is a boolean mask that marks the missing values. This is analogous to the np.isnan function for arrays, which we learned about earlier (see Detecting “No Data” (np.isnan)).
For example, edu.isna() returns a boolean DataFrame, of the same shape as edu, with True marking “No Data” values:
edu.isna()
| name | type | address | old_name | lat | lon | |
|---|---|---|---|---|---|---|
| 0 | False | False | False | True | False | False |
| 1 | False | False | False | True | False | False |
| 2 | False | False | False | True | False | False |
| 3 | False | False | False | True | False | False |
| 4 | False | False | False | True | False | False |
| ... | ... | ... | ... | ... | ... | ... |
| 484 | False | False | False | True | False | False |
| 485 | False | False | False | True | False | False |
| 486 | False | False | False | True | False | False |
| 487 | False | False | False | False | False | False |
| 488 | False | False | False | False | False | False |
489 rows × 6 columns
Similarly, the .isna() method applied on a Series returns a new boolean Series where “No Data” values are marked with True:
edu['old_name'].isna()
0 True
1 True
2 True
3 True
4 True
...
484 True
485 True
486 True
487 False
488 False
Name: old_name, Length: 489, dtype: bool
The ~ operator (see numpy conditional operators) reverses a boolean Series. This is very useful in the context of “No Data” masks, since it can reverse the mask so that it points at non-missing values.
For example, here is how we can get the reverse of edu.isna():
~edu.isna()
| name | type | address | old_name | lat | lon | |
|---|---|---|---|---|---|---|
| 0 | True | True | True | False | True | True |
| 1 | True | True | True | False | True | True |
| 2 | True | True | True | False | True | True |
| 3 | True | True | True | False | True | True |
| 4 | True | True | True | False | True | True |
| ... | ... | ... | ... | ... | ... | ... |
| 484 | True | True | True | False | True | True |
| 485 | True | True | True | False | True | True |
| 486 | True | True | True | False | True | True |
| 487 | True | True | True | True | True | True |
| 488 | True | True | True | True | True | True |
489 rows × 6 columns
and here is how we can get the reverse of edu['old_name'].isna():
~edu['old_name'].isna()
0 False
1 False
2 False
3 False
4 False
...
484 False
485 False
486 False
487 True
488 True
Name: old_name, Length: 489, dtype: bool
Another possible use of ~ and .isna is to extract the non-missing values from a series. Here is an example:
edu['old_name'][~edu['old_name'].isna()]
6 רד"ק
7 השלושה
10 ברנר
13 רקפת
19 מיתרים
...
365 עובדי מדינה
400 גור אריה
415 תבור
487 מיתרים
488 מיתרים
Name: old_name, Length: 67, dtype: object
However, there is a dedicated method called .dropna to do that with shorter code (see Removing with .dropna):
edu['old_name'].dropna()
6 רד"ק
7 השלושה
10 ברנר
13 רקפת
19 מיתרים
...
365 עובדי מדינה
400 גור אריה
415 תבור
487 מיתרים
488 מיתרים
Name: old_name, Length: 67, dtype: object
Similarly, we can extract DataFrame rows where a particular column is non-missing. For example, here we have those edu rows that have a non-missing 'species' value:
edu[~edu['old_name'].isna()]
| name | type | address | old_name | lat | lon | |
|---|---|---|---|---|---|---|
| 6 | מאדים | גן ילדים | רד"ק שכ יא | רד"ק | 31.256963 | 34.772220 |
| 7 | אדמה | גן ילדים | רח טבריה שכ ט | השלושה | 31.247157 | 34.776405 |
| 10 | חלילית | גן ילדים | ברנר שכ א | ברנר | 31.248934 | 34.793113 |
| 13 | חרצית | גן ילדים | מבצע ארז שכ ו | רקפת | 31.265425 | 34.782446 |
| 19 | איילה | גן ילדים | וינגיט שכ ג | מיתרים | 31.256078 | 34.803588 |
| ... | ... | ... | ... | ... | ... | ... |
| 365 | נבל | גן ילדים | NaN | עובדי מדינה | 31.248385 | 34.786168 |
| 400 | נווה | גן ילדים | המר 3 | גור אריה | 31.266327 | 34.810049 |
| 415 | ארבל | גן ילדים | מודעי | תבור | 31.266514 | 34.809769 |
| 487 | שמים | גן ילדים | וינגיט שכ ג | מיתרים | 31.254752 | 34.803860 |
| 488 | תכלת | גן ילדים | וינגיט שכ ג | מיתרים | 31.254470 | 34.804140 |
67 rows × 6 columns
Note
Instead of reversing the output of .isna with the ~ operator, another option is to use the inverse method .notna, which returns the non-missing mask.
Finally, it is often useful to combine the .isna method with .any to find out if a Series contains at least one missing value:
edu['old_name'].isna().any()
np.True_
edu['name'].isna().any()
np.False_
or to find out which columns contain at least one missing value:
edu.isna().any()
name False
type False
address True
old_name True
lat False
lon False
dtype: bool
We can also use .sum to find out how many missing value are there in each column:
edu.isna().sum()
name 0
type 0
address 86
old_name 422
lat 0
lon 0
dtype: int64
or .mean to find the proportion of missing values in each column:
edu.isna().mean()
name 0.000000
type 0.000000
address 0.175869
old_name 0.862986
lat 0.000000
lon 0.000000
dtype: float64
Operators with missing values#
When applying a method such as .sum, .mean, .min, or .max (or the respective np.* function) on a Series, the “No Data” values are ignored. That is, the result is based only on the non-missing values. For example:
edu.loc[~edu['old_name'].isna(), 'old_name1'] = 1
edu
| name | type | address | old_name | lat | lon | old_name1 | |
|---|---|---|---|---|---|---|---|
| 0 | דוגית | גן ילדים | משה סתוי נ.זאב | NaN | 31.240226 | 34.767153 | NaN |
| 1 | שומרון | גן ילדים | שומרון שכ לון | NaN | 31.246547 | 34.765438 | NaN |
| 2 | שרת | גן ילדים | משה שרת 13 | NaN | 31.256681 | 34.766891 | NaN |
| 3 | חופית | גן ילדים | ניב דוד נ.זאב | NaN | 31.238660 | 34.770356 | NaN |
| 4 | רביבים | גן ילדים | מבצע משה נ.זאב | NaN | 31.237117 | 34.779331 | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 484 | ברעם | גן ילדים | נחל משמר | NaN | 31.233890 | 34.818100 | NaN |
| 485 | שילה | גן ילדים | נחל יהל 3 | NaN | 31.232486 | 34.821510 | NaN |
| 486 | קדם | גן ילדים | נחל יהל 3 | NaN | 31.232557 | 34.821390 | NaN |
| 487 | שמים | גן ילדים | וינגיט שכ ג | מיתרים | 31.254752 | 34.803860 | 1.0 |
| 488 | תכלת | גן ילדים | וינגיט שכ ג | מיתרים | 31.254470 | 34.804140 | 1.0 |
489 rows × 7 columns
edu['old_name1'].sum()
np.float64(67.0)
Note that this behavior is specific to pandas, and unlike what we have seen with numpy arrays, where these opertions return “No Data” if at least one of the values in the array is “No Data” (see Operations with “No Data”):
edu['old_name1'].to_numpy().sum()
np.float64(nan)
Element-by-element operations, however, expectedly result in “No Data” when applied to “No Data” elements, since the result cannot be computed at all. For example:
edu['old_name1'] * 2
0 NaN
1 NaN
2 NaN
3 NaN
4 NaN
...
484 NaN
485 NaN
486 NaN
487 2.0
488 2.0
Name: old_name1, Length: 489, dtype: float64
Removing with .dropna#
The .dropna method can be used to remove missing values from a DataFrame (or a Series). The default is to drop all rows that have at least one missing value, i.e., keeping only complete observations. For example:
edu.dropna()
| name | type | address | old_name | lat | lon | old_name1 | |
|---|---|---|---|---|---|---|---|
| 6 | מאדים | גן ילדים | רד"ק שכ יא | רד"ק | 31.256963 | 34.772220 | 1.0 |
| 7 | אדמה | גן ילדים | רח טבריה שכ ט | השלושה | 31.247157 | 34.776405 | 1.0 |
| 10 | חלילית | גן ילדים | ברנר שכ א | ברנר | 31.248934 | 34.793113 | 1.0 |
| 13 | חרצית | גן ילדים | מבצע ארז שכ ו | רקפת | 31.265425 | 34.782446 | 1.0 |
| 19 | איילה | גן ילדים | וינגיט שכ ג | מיתרים | 31.256078 | 34.803588 | 1.0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 363 | אוריה | גן ילדים | בי"ס מוריה שכ א | נריה | 31.246930 | 34.792426 | 1.0 |
| 400 | נווה | גן ילדים | המר 3 | גור אריה | 31.266327 | 34.810049 | 1.0 |
| 415 | ארבל | גן ילדים | מודעי | תבור | 31.266514 | 34.809769 | 1.0 |
| 487 | שמים | גן ילדים | וינגיט שכ ג | מיתרים | 31.254752 | 34.803860 | 1.0 |
| 488 | תכלת | גן ילדים | וינגיט שכ ג | מיתרים | 31.254470 | 34.804140 | 1.0 |
59 rows × 7 columns
Note
There are other parameters of .dropna for dropping columns instead of rows, or dropping rows or column using a threshold of “acceptable” minimum number of non-missing values. See the documentation for details.
Replacing with .fillna#
Missing values can be replaced using the .fillna method. The parameter specifies which new value should be placed instead of the “No Data” values. For example, here is how we can replace the “No Data” values in the 'sigalon' column of edu with 0:
edu['old_name1'].fillna(0)
0 0.0
1 0.0
2 0.0
3 0.0
4 0.0
...
484 0.0
485 0.0
486 0.0
487 1.0
488 1.0
Name: old_name1, Length: 489, dtype: float64
or assign the result back to the edu table:
edu['old_name1'] = edu['old_name1'].fillna(0)
edu
| name | type | address | old_name | lat | lon | old_name1 | |
|---|---|---|---|---|---|---|---|
| 0 | דוגית | גן ילדים | משה סתוי נ.זאב | NaN | 31.240226 | 34.767153 | 0.0 |
| 1 | שומרון | גן ילדים | שומרון שכ לון | NaN | 31.246547 | 34.765438 | 0.0 |
| 2 | שרת | גן ילדים | משה שרת 13 | NaN | 31.256681 | 34.766891 | 0.0 |
| 3 | חופית | גן ילדים | ניב דוד נ.זאב | NaN | 31.238660 | 34.770356 | 0.0 |
| 4 | רביבים | גן ילדים | מבצע משה נ.זאב | NaN | 31.237117 | 34.779331 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 484 | ברעם | גן ילדים | נחל משמר | NaN | 31.233890 | 34.818100 | 0.0 |
| 485 | שילה | גן ילדים | נחל יהל 3 | NaN | 31.232486 | 34.821510 | 0.0 |
| 486 | קדם | גן ילדים | נחל יהל 3 | NaN | 31.232557 | 34.821390 | 0.0 |
| 487 | שמים | גן ילדים | וינגיט שכ ג | מיתרים | 31.254752 | 34.803860 | 1.0 |
| 488 | תכלת | גן ילדים | וינגיט שכ ג | מיתרים | 31.254470 | 34.804140 | 1.0 |
489 rows × 7 columns
Views and copies (pandas)#
One more thing we need to be aware when working with pandas is the distinction between views and copies. The pandas package (and geopandas which extends it, see Vector layers (geopandas)) behaves similarly to numpy with respect to copies (see Views and copies (numpy)). We must be aware of whether we are creating a view or a copy of a DataFrame, to avoid unexpected results.
Here is a small demonstration, very similar to the one we did with numpy arrays (see Views and copies (numpy)). Suppose that we create a copy of dat, named dat2:
dat2 = dat
Next, we modify dat2, assigning a new value such as 9999 into a particular cell (e.g., 2nd row, 2nd column):
dat2.iloc[1, 1] = 9999
The printout of the table top-left “corner” demonstrates the value was actually changed:
dat2.iloc[:4, :4]
| Year | Glob | NHem | SHem | |
|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 |
| 1 | 1881 | 9999.00 | -0.18 | 0.00 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 |
What may be surprising, again, is that the original table dat has also changed. We thus demonstrated that dat2 is a reference to dat, rather than an independent copy:
dat.iloc[:4, :4]
| Year | Glob | NHem | SHem | |
|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 |
| 1 | 1881 | 9999.00 | -0.18 | 0.00 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 |
To create an independent copy, so that its modifications are not reflected in the original, we need to explicitly use the .copy method, as in:
dat2 = dat.copy()
Note
The behavior described in this section can be disabled with pd.options.mode.copy_on_write=True, and will be disabled by default in pandas version 3.0. When disabled, every pandas operation will create an independent copy, and, for example, it would be impossible to update two objects at once as done above.
Let us read the CSV file again to get back to the original values:
dat = pd.read_csv('data/ZonAnn.Ts+dSST.csv')
Writing DataFrame to file#
A DataFrame can be exported to several formats, such as CSV or Excel, using the appropriate method. For example, exporting to CSV is done using the .to_csv method.
Here is how we can export the DataFrame named stations, which we created in the beginning of the chapter (see Creating a DataFrame), to a CSV file named stations.csv in the output directory. The additional index=False argument specifies that we do not want to export the index values, which is usually the case:
stations.to_csv('output/stations.csv', index=False)
Note that the file is exported to a sub-directory named output (which must already exist!).
The way the resulting file stations.csv is displayed in a spreadsheet program, such as Microsoft Excel or LibreOffice Calc, is shown in Fig. 28.
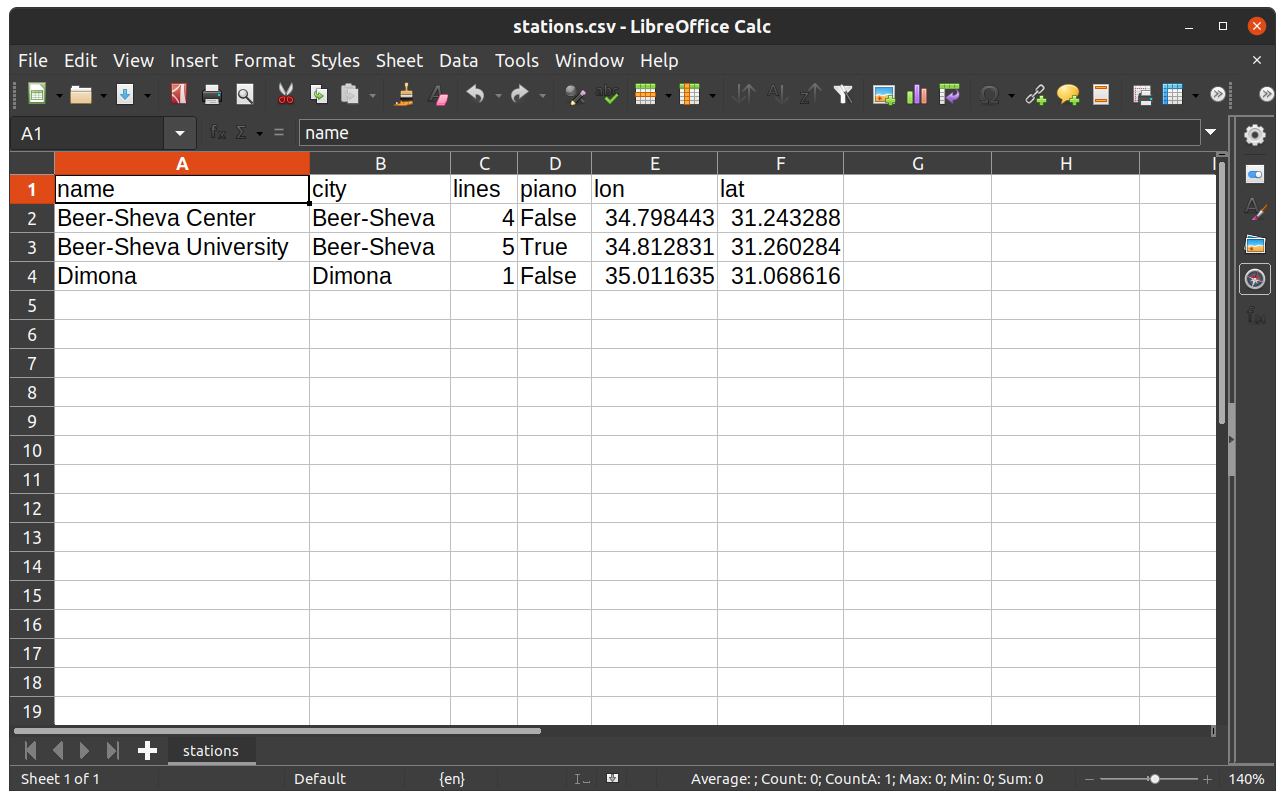
Fig. 28 The DataFrame named stations when exported to CSV file and opened in LibreOffice Calc#
More exercises#
Exercise 05-g
Import the file
'education.csv'into aDataFrameRename the columns as shown in Missing data in pandas
How many observations with non-missing
'old_name'are there?How many observations with non-missing
'address'and non-missing'old_name'are there?How many unique instytution types are there?
Create a
listof the unique types and print it
Exercise solutions#
Exercise 05-d#
import pandas as pd
dat = pd.read_csv('data/ZonAnn.Ts+dSST.csv')
dat['Year'].iloc[dat.shape[0]-1]
np.int64(2023)
Exercise 05-g#
import pandas as pd
edu = pd.read_csv('data/education.csv')
edu = edu.rename(columns={
'Institutio': 'name',
'ADDRESS': 'address',
'oldName': 'old_name'
})
(~edu['old_name'].isna()).sum()
np.int64(67)
(~edu['address'].isna() & ~edu['old_name'].isna()).sum()
np.int64(59)
edu['type'].nunique()
5
edu['type'].unique().tolist()
['גן ילדים', 'מרכז מדעים', 'חווה חקלאי', 'על יסודי', 'בית ספר']