Table reshaping and joins#
Last updated: 2025-01-15 17:42:48
Introduction#
In the previous chapter (see Tables (pandas)), we introduced the pandas package, and covered the basic operations with tables using pandas, such as importing and exporting, subsetting, renaming columns, sorting, and calculating new columns. In practice, we often need to do more than that. For example, we may need to aggregate, or to reshape, tables into a different form, or to combine numerous tables into one. In this chapter, we cover some of the common table operations which involve modifying table structure or combining multiple tables.
When working with data, it is often necessary to combine data from multiple sources, and/or reshape data into the right form that a particular function accepts:
Combining may simply involve “pasting”, or concatenating, two tables one on top of the other, or side-by-side (see Concatenation).
In more complex cases, combining involves a table join, where the information from two tables is combined according to one or more “key” columns which are common to both tables (see Joining tables).
Moreover, we may need to summarize the values of a table according to a particular grouping, which is known as aggregation (see Aggregation).
A special type of aggregation is applying a function across each row, or each column, of a table (Row/col-wise operations).
In this chapter, we learn about all of those common methods for combining and reshaping tables. By the end of the chapter, you will have the ability to not just import and transform tables in Python (see Tables (pandas)), but also to reshape and combine tables to the right form, which facilitates further analysis and insight from the data in those tables.
Sample data#
First, let’s recreate the small table named stations which we used in the previous chapter (see Creating a DataFrame):
import numpy as np
import pandas as pd
name = pd.Series(['Beer-Sheva Center', 'Beer-Sheva University', 'Dimona'])
city = pd.Series(['Beer-Sheva', 'Beer-Sheva', 'Dimona'])
lines = pd.Series([4, 5, 1])
piano = pd.Series([False, True, False])
lon = pd.Series([34.798443, 34.812831, 35.011635])
lat = pd.Series([31.243288, 31.260284, 31.068616])
d = {'name': name, 'city': city, 'lines': lines, 'piano': piano, 'lon': lon, 'lat': lat}
stations = pd.DataFrame(d)
stations
| name | city | lines | piano | lon | lat | |
|---|---|---|---|---|---|---|
| 0 | Beer-Sheva Center | Beer-Sheva | 4 | False | 34.798443 | 31.243288 |
| 1 | Beer-Sheva University | Beer-Sheva | 5 | True | 34.812831 | 31.260284 |
| 2 | Dimona | Dimona | 1 | False | 35.011635 | 31.068616 |
We will also load the sample climatic data which we are also familiar with from the previous chapter (Reading from file), into a DataFrame named dat:
dat = pd.read_csv('data/ZonAnn.Ts+dSST.csv')
dat
| Year | Glob | NHem | SHem | 24N-90N | ... | EQU-24N | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 | -0.38 | ... | -0.14 | -0.11 | -0.04 | 0.05 | 0.67 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 | -0.36 | ... | 0.11 | 0.10 | -0.06 | -0.07 | 0.60 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 | -0.31 | ... | -0.04 | -0.05 | 0.01 | 0.04 | 0.63 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 | -0.34 | ... | -0.17 | -0.16 | -0.04 | 0.07 | 0.50 |
| 4 | 1884 | -0.28 | -0.42 | -0.15 | -0.61 | ... | -0.12 | -0.17 | -0.19 | -0.02 | 0.65 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 2019 | 0.98 | 1.20 | 0.75 | 1.42 | ... | 0.90 | 0.90 | 0.75 | 0.39 | 0.83 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 | 1.67 | ... | 0.88 | 0.84 | 0.58 | 0.39 | 0.89 |
| 141 | 2021 | 0.85 | 1.14 | 0.56 | 1.42 | ... | 0.72 | 0.60 | 0.72 | 0.32 | 0.30 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 | 1.52 | ... | 0.62 | 0.51 | 0.79 | 0.38 | 1.09 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 | 1.78 | ... | 1.07 | 1.04 | 0.91 | 0.44 | 0.63 |
144 rows × 15 columns
Concatenation#
Overview#
The most basic method for combining several tables into one is concatenation, which essentially means binding tables one on top of another, or side-by-side:
Concatenation by row is usually desired when the two (or more) tables share exactly the same columns. For example, combining several tables reflecting the same data (e.g., measured on different dates, or for different subjects) involves concatenation by row.
Concatenation by column is useful when we need to attach new variables for the same observations we already have. For example, we may want to combine several different types of observations which are split across separate files.
In pandas, concatenation can be done using pd.concat. The pd.concat function accepts a list of tables, which are combined:
By row when using
axis=0(the default)By column when using
axis=1
Concatenation by row#
To demonstrate concatenation by row, let’s create two subsets x and y of stations, with the 1st row and the 2nd-3rd rows, respectively:
x = stations.iloc[[0], :] ## 1st row
x
| name | city | lines | piano | lon | lat | |
|---|---|---|---|---|---|---|
| 0 | Beer-Sheva Center | Beer-Sheva | 4 | False | 34.798443 | 31.243288 |
y = stations.iloc[[1, 2], :] ## 2nd and 3rd rows
y
| name | city | lines | piano | lon | lat | |
|---|---|---|---|---|---|---|
| 1 | Beer-Sheva University | Beer-Sheva | 5 | True | 34.812831 | 31.260284 |
| 2 | Dimona | Dimona | 1 | False | 35.011635 | 31.068616 |
The expression pd.concat([x,y]) can then be used to concatenate x and y, by row, re-creating the complete table stations:
pd.concat([x, y])
| name | city | lines | piano | lon | lat | |
|---|---|---|---|---|---|---|
| 0 | Beer-Sheva Center | Beer-Sheva | 4 | False | 34.798443 | 31.243288 |
| 1 | Beer-Sheva University | Beer-Sheva | 5 | True | 34.812831 | 31.260284 |
| 2 | Dimona | Dimona | 1 | False | 35.011635 | 31.068616 |
Note that pd.concat accepts a list of DataFrames to concatenate, such as [x,y]. In this example the list is of length 2, but in general it can be of any length.
Concatenation by column#
To demonstrate concatenation by column, let’s now split stations to subsets x and y by column. For example, the x table is going to contain one column 'name':
x = stations[['name']]
x
| name | |
|---|---|
| 0 | Beer-Sheva Center |
| 1 | Beer-Sheva University |
| 2 | Dimona |
and the y table is going to contain two columns 'lon' and 'lat':
y = stations[['lon', 'lat']]
y
| lon | lat | |
|---|---|---|
| 0 | 34.798443 | 31.243288 |
| 1 | 34.812831 | 31.260284 |
| 2 | 35.011635 | 31.068616 |
To concatenate by column, we use pd.concat with the axis=1 argument. This concatenates the tables x and y side-by-side:
pd.concat([x, y], axis=1)
| name | lon | lat | |
|---|---|---|---|
| 0 | Beer-Sheva Center | 34.798443 | 31.243288 |
| 1 | Beer-Sheva University | 34.812831 | 31.260284 |
| 2 | Dimona | 35.011635 | 31.068616 |
Alignment by index#
Note that concatenation—like many other operations in pandas—operates based on the index, rather than position. This is not a problem when concatenating by row, where columns typically need to be aligned by their index, i.e., column name (see Concatenation by row). When concatenating by column, however, we need to pay attention to the index: is it meaningful, and we want to use it when aligning rows? Or is it arbitarary, and we want to reset it before concatenating?
For example, here is what happens in concatenation by column, when the index is not aligned (see Setting Series index):
y.index = [1,2,3]
pd.concat([x, y], axis=1)
| name | lon | lat | |
|---|---|---|---|
| 0 | Beer-Sheva Center | NaN | NaN |
| 1 | Beer-Sheva University | 34.798443 | 31.243288 |
| 2 | Dimona | 34.812831 | 31.260284 |
| 3 | NaN | 35.011635 | 31.068616 |
To make sure we concatenate based on position, we may reset the index (using .reset_index(drop=True)) on both tables before concatenation:
y = y.reset_index(drop=True)
pd.concat([x, y], axis=1)
| name | lon | lat | |
|---|---|---|---|
| 0 | Beer-Sheva Center | 34.798443 | 31.243288 |
| 1 | Beer-Sheva University | 34.812831 | 31.260284 |
| 2 | Dimona | 35.011635 | 31.068616 |
Row/col-wise operations#
Built-in methods#
pandas defines summary methods which can be used to calculate row-wise and column-wise summaries of DataFrames. The pandas methods are generally similar to the numpy 2D summary functions and methods (see Summaries per dimension (2D)) which we learned about earlier, but there are several important differences to keep in mind:
pandasonly has methods, not functions, i.e., there is a.meanmethod but there is nopd.meanfunction—unlikenumpy, which in many cases has both, e.g., both.meanandnp.mean(see Table 13)The default
axisis different: inpandasthe default is column-wise summary (axis=0), while innumpythe default is “global” summary (axis=None) (see Global summaries)pandasmethods exclude “No Data” by default (see Operators with missing values), unless we explicitly specifyskipna=False—unlikenumpywhich by default includes “No Data”, unless using the specific “No Data”-safe functions such asnp.nanmean(see Operations with “No Data”)
Commonly used pandas summary methods are listed in Table 14.
Operation |
Method |
|---|---|
Sum |
|
Minimum |
|
Maximum |
|
Mean |
|
Median |
|
Count (of non-missing values) |
|
Number of unique values |
|
Index of (first) minimum |
|
Index of (first) maximum |
|
Is at least one element |
|
Are all elements |
For the next few examples, let’s take the regional temperature anomaly time series into a separate DataFrame named regions:
cols = [
'90S-64S',
'64S-44S',
'44S-24S',
'24S-EQU',
'EQU-24N',
'24N-44N',
'44N-64N',
'64N-90N'
]
regions = dat[cols]
regions
| 90S-64S | 64S-44S | 44S-24S | 24S-EQU | EQU-24N | 24N-44N | 44N-64N | 64N-90N | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0.67 | 0.05 | -0.04 | -0.11 | -0.14 | -0.26 | -0.52 | -0.80 |
| 1 | 0.60 | -0.07 | -0.06 | 0.10 | 0.11 | -0.19 | -0.50 | -0.93 |
| 2 | 0.63 | 0.04 | 0.01 | -0.05 | -0.04 | -0.13 | -0.31 | -1.41 |
| 3 | 0.50 | 0.07 | -0.04 | -0.16 | -0.17 | -0.24 | -0.58 | -0.18 |
| 4 | 0.65 | -0.02 | -0.19 | -0.17 | -0.12 | -0.45 | -0.66 | -1.30 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 0.83 | 0.39 | 0.75 | 0.90 | 0.90 | 0.99 | 1.43 | 2.71 |
| 140 | 0.89 | 0.39 | 0.58 | 0.84 | 0.88 | 1.19 | 1.81 | 2.88 |
| 141 | 0.30 | 0.32 | 0.72 | 0.60 | 0.72 | 1.26 | 1.35 | 2.05 |
| 142 | 1.09 | 0.38 | 0.79 | 0.51 | 0.62 | 1.27 | 1.50 | 2.34 |
| 143 | 0.63 | 0.44 | 0.91 | 1.04 | 1.07 | 1.47 | 1.87 | 2.58 |
144 rows × 8 columns
Using the .mean method combined with axis=0 (i.e., the function is applied on each column), we can calculate the mean temperature anomaly in each region:
regions.mean(axis=0)
90S-64S -0.074028
64S-44S -0.053681
44S-24S 0.047917
24S-EQU 0.083056
EQU-24N 0.066181
24N-44N 0.052500
44N-64N 0.142500
64N-90N 0.273333
dtype: float64
Note that axis=0 is the default, so we can also get the same result without specifying it (unlike numpy where the default is global summary, i.e., axis=None; see Global summaries):
regions.mean()
90S-64S -0.074028
64S-44S -0.053681
44S-24S 0.047917
24S-EQU 0.083056
EQU-24N 0.066181
24N-44N 0.052500
44N-64N 0.142500
64N-90N 0.273333
dtype: float64
Exercise 06-a
Calculate a
listof (the first) years when the maximum temperature was observed per region (answer: [1996, 1985, 2023, 2016, 2023, 2023, 2023, 2016])
Conversely, using axis=1, we can calculate row means, i.e., the average temperature anomaly for each year across all regions:
regions.mean(axis=1)
0 -0.14375
1 -0.11750
2 -0.15750
3 -0.10000
4 -0.28250
...
139 1.11250
140 1.18250
141 0.91500
142 1.06250
143 1.25125
Length: 144, dtype: float64
If necessary, the result can be assigned into a new column (see Creating new columns), hereby named mean, as follows:
dat['mean'] = regions.mean(axis=1)
dat
| Year | Glob | NHem | SHem | 24N-90N | ... | 24S-EQU | 44S-24S | 64S-44S | 90S-64S | mean | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1880 | -0.17 | -0.28 | -0.05 | -0.38 | ... | -0.11 | -0.04 | 0.05 | 0.67 | -0.14375 |
| 1 | 1881 | -0.09 | -0.18 | 0.00 | -0.36 | ... | 0.10 | -0.06 | -0.07 | 0.60 | -0.11750 |
| 2 | 1882 | -0.11 | -0.21 | -0.01 | -0.31 | ... | -0.05 | 0.01 | 0.04 | 0.63 | -0.15750 |
| 3 | 1883 | -0.17 | -0.28 | -0.07 | -0.34 | ... | -0.16 | -0.04 | 0.07 | 0.50 | -0.10000 |
| 4 | 1884 | -0.28 | -0.42 | -0.15 | -0.61 | ... | -0.17 | -0.19 | -0.02 | 0.65 | -0.28250 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | 2019 | 0.98 | 1.20 | 0.75 | 1.42 | ... | 0.90 | 0.75 | 0.39 | 0.83 | 1.11250 |
| 140 | 2020 | 1.01 | 1.35 | 0.68 | 1.67 | ... | 0.84 | 0.58 | 0.39 | 0.89 | 1.18250 |
| 141 | 2021 | 0.85 | 1.14 | 0.56 | 1.42 | ... | 0.60 | 0.72 | 0.32 | 0.30 | 0.91500 |
| 142 | 2022 | 0.89 | 1.16 | 0.62 | 1.52 | ... | 0.51 | 0.79 | 0.38 | 1.09 | 1.06250 |
| 143 | 2023 | 1.17 | 1.49 | 0.85 | 1.78 | ... | 1.04 | 0.91 | 0.44 | 0.63 | 1.25125 |
144 rows × 16 columns
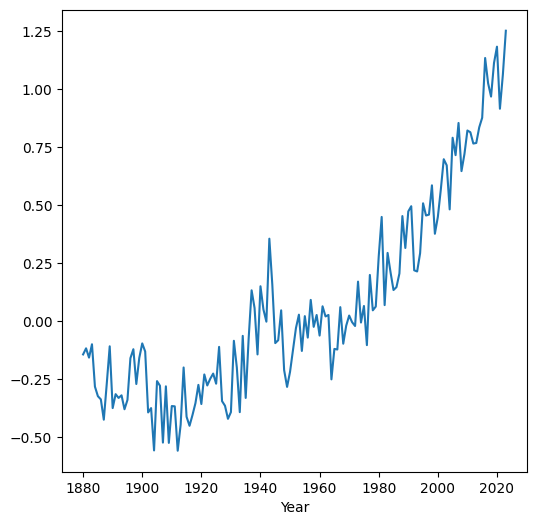
Here is a visualization (see Line plots (pandas)) of the mean time series we just calculated:
dat.set_index('Year')['mean'].plot();
apply and custom functions#
In case we need to apply a custom function on each row, or column, we can use the .apply method combined with a lambda function (see Lambda functions). For example, here is an alternative way to calculate row-wise means, using the .apply method and a lambda function. The result is the same as regions.mean() (see Built-in methods):
regions.apply(lambda x: x.mean())
90S-64S -0.074028
64S-44S -0.053681
44S-24S 0.047917
24S-EQU 0.083056
EQU-24N 0.066181
24N-44N 0.052500
44N-64N 0.142500
64N-90N 0.273333
dtype: float64
The advantage is that we can use any custom function definition. For example, the following expression calculates whether each region had temperature anomaly above 1 degree, in at least one of the 140 years:
regions.apply(lambda x: (x > 1).any())
90S-64S True
64S-44S False
44S-24S False
24S-EQU True
EQU-24N True
24N-44N True
44N-64N True
64N-90N True
dtype: bool
Similarly to the built-in summary functions (see Built-in methods), .apply uses the axis argument, with axis=0 specifying column-wise summaries as shown in the last two examples (the default), or axis=1 for row-wise summaries.
Exercise 06-b
Function
np.polyfitcan be used to calculate the slope of linear regression between two columns. For example, using the code section shown below, you can find out that the linear trend of temperature in the'44S-24S'region was an increasing one, at0.0066degrees per year.Adapt the code into to the form of a function named
slopewhich, given twoSeriesobjectsxandy, calculates the linear slope. For example,f(dat['Year'],regions['44S-24S'])should return0.0066.Use the
slopefunction you defined within the.applymethod, to calculate the slope in all columns ofregionsat once.Hint: you need to keep the
xargument ofslopefixed atdat['Year'], while the various columns ofregionsare passed toy. To do that, write another function (or use a lambda function) that “wraps”slopeand runs it with just theyparameter (keepingxfixed).
x = dat['Year']
y = regions['44S-24S']
np.polyfit(x, y, 1)[0]
Joining tables#
Overview#
One of the most common operations when working with data is a table join. In a table join, two tables are combined into one, based on one or more common column(s). For example, when compiling a dataset about different cities, we may collect data on different aspects of each city (population size, socio-economic status, built area, etc.) from different sources. To work with these data together, the various tables need to be joined into one. The common column, in this case, would be the city name, or city ID.
When working with pandas, tables can be joined, based on one or more common column, using the pd.merge function. pd.merge has the following important parameters:
left—The first (“left”) tableright—The second (“right”) tablehow—The type of join, one of:'left','right','outer','inner'(the default),'cross'on—The name(s) of the common column(s) where identical values are considered a “match”
The type of join (the how parameter) basically determines what should be done with non-matching records. The options are 'left', 'right', 'outer', 'inner' (the default), and 'cross'. The meaning of the join types is the same as in SQL[1], and will be demonstrated through examples below.
Stations example#
For a first small example, let’s get back to the stations table:
stations
| name | city | lines | piano | lon | lat | |
|---|---|---|---|---|---|---|
| 0 | Beer-Sheva Center | Beer-Sheva | 4 | False | 34.798443 | 31.243288 |
| 1 | Beer-Sheva University | Beer-Sheva | 5 | True | 34.812831 | 31.260284 |
| 2 | Dimona | Dimona | 1 | False | 35.011635 | 31.068616 |
Suppose we have another small table named status which describes the status ('Closed' or 'Open') of select stations:
name = pd.Series(['Beer-Sheva Center', 'Beer-Sheva University', 'Tel-Aviv University'])
status = pd.Series(['Closed', 'Open', 'Open'])
status = pd.DataFrame({'name': name, 'status': status})
status
| name | status | |
|---|---|---|
| 0 | Beer-Sheva Center | Closed |
| 1 | Beer-Sheva University | Open |
| 2 | Tel-Aviv University | Open |
The stations and status can be joined as follows:
pd.merge(stations, status)
| name | city | lines | piano | lon | lat | status | |
|---|---|---|---|---|---|---|---|
| 0 | Beer-Sheva Center | Beer-Sheva | 4 | False | 34.798443 | 31.243288 | Closed |
| 1 | Beer-Sheva University | Beer-Sheva | 5 | True | 34.812831 | 31.260284 | Open |
This implies the default, on being equal to the column(s) sharing the same name in both tables (on='name', in this case), and an 'inner' join (i.e., how='inner'). As you can see, an inner join returns just the matching rows. It is better to be explicit, for clarity:
pd.merge(stations, status, on='name', how='inner')
| name | city | lines | piano | lon | lat | status | |
|---|---|---|---|---|---|---|---|
| 0 | Beer-Sheva Center | Beer-Sheva | 4 | False | 34.798443 | 31.243288 | Closed |
| 1 | Beer-Sheva University | Beer-Sheva | 5 | True | 34.812831 | 31.260284 | Open |
Unlike an 'inner' join, a 'left' join returns all records from the first (“left”) table, even those that have no match in the second (“right”) table. For example, 'Dimona' station does not appear in the status table, but it is still returned in the result (with status set to np.nan):
pd.merge(stations, status, on='name', how='left')
| name | city | lines | piano | lon | lat | status | |
|---|---|---|---|---|---|---|---|
| 0 | Beer-Sheva Center | Beer-Sheva | 4 | False | 34.798443 | 31.243288 | Closed |
| 1 | Beer-Sheva University | Beer-Sheva | 5 | True | 34.812831 | 31.260284 | Open |
| 2 | Dimona | Dimona | 1 | False | 35.011635 | 31.068616 | NaN |
A 'right' join is the opposite of a 'left' join, i.e., it returns all records from the “right” table, even those that have no match in the “left” table. This time, values absent in the “left” table are set to np.nan:
pd.merge(stations, status, on='name', how='right')
| name | city | lines | piano | lon | lat | status | |
|---|---|---|---|---|---|---|---|
| 0 | Beer-Sheva Center | Beer-Sheva | 4.0 | False | 34.798443 | 31.243288 | Closed |
| 1 | Beer-Sheva University | Beer-Sheva | 5.0 | True | 34.812831 | 31.260284 | Open |
| 2 | Tel-Aviv University | NaN | NaN | NaN | NaN | NaN | Open |
Finally, an 'outer' (also known as “full”) join returns all record from both tables, regardless of whether they have a matching record in the other table. Values that have no match in both the “left” and “right” tables are set to np.nan:
pd.merge(stations, status, on='name', how='outer')
| name | city | lines | piano | lon | lat | status | |
|---|---|---|---|---|---|---|---|
| 0 | Beer-Sheva Center | Beer-Sheva | 4.0 | False | 34.798443 | 31.243288 | Closed |
| 1 | Beer-Sheva University | Beer-Sheva | 5.0 | True | 34.812831 | 31.260284 | Open |
| 2 | Dimona | Dimona | 1.0 | False | 35.011635 | 31.068616 | NaN |
| 3 | Tel-Aviv University | NaN | NaN | NaN | NaN | NaN | Open |
When working with data, we often have one main, or “template”, table we are working with (“left”) and we are interested to join data coming from another external (“right”) table to it. In this scenario we usually prefer not to lose any records from the “left” table. Even if some of the records have no match and get “No Data” values (namely, np.nan), we usually prefer to keep those records and deal with the missing values as necessary, rather than lose the records altogether. Therefore, a 'left' join is often most useful in data analysis. For example, in the above situation a 'left' join makes sense if our focus is on the station properties given in stations, so even though the status of 'Dimona' is unknown—we prefer to keep the 'Dimona' station record.
What is GTFS?#
In the next examples, we demonstrate table joins on a realistic large dataset, namely a GTFS public transport dataset. Before we go into the example, we need to be familiar with the GTFS format and structure.
The General Transit Feed Specification (GTFS) is a standard format for describing public transport routes and timetables. GTFS is typically published by government transport agencies, periodically. For example, the GTFS dataset provided as part of the book’s sample data was downloaded from Israel ministry of transport GTFS website on '2023-09-30'. GTFS data are used by both commercial companies, such as for routing in Google Maps, and in open-source tools.
Technically, a GTFS dataset is a collection of several CSV files (even though their file extension is '.txt'), of specific designation and structure, linked via common columns, or keys. In the examples, we are going to use only the six most important files of the GTFS dataset (Table 15). Recall that we already met one of the GTFS files, namely 'stops.txt', earlier, in an example of text file processing through for loops (see File object and reading lines).
Name |
Contents |
Keys |
|---|---|---|
|
Agencies |
|
|
Routes |
|
|
Trips |
|
|
Stop times |
|
|
Stops |
|
|
Shapes |
|
Let’s see what the GTFS tables look like, what is their purpose, and how are they related with one another. To make things easier, we immediately subset the most important columns, which are also the ones we use in subsequent examples.
We start with 'agency.txt'. This file lists the public transport agencies. Each row represents one agency, identified by 'agency_id' (note that we are using usecols to read just the necessary columns from the CSV file):
cols = ['agency_id', 'agency_name']
pd.read_csv('data/gtfs/agency.txt', usecols=cols)
| agency_id | agency_name | |
|---|---|---|
| 0 | 2 | רכבת ישראל |
| 1 | 3 | אגד |
| 2 | 4 | אלקטרה אפיקים תחבורה |
| 3 | 5 | דן |
| 4 | 6 | ש.א.מ |
| ... | ... | ... |
| 31 | 51 | ירושלים-צור באהר איחוד |
| 32 | 91 | מוניות מטרו קו |
| 33 | 93 | מוניות מאיה יצחק שדה |
| 34 | 97 | אודליה מוניות בעמ |
| 35 | 98 | מוניות רב קווית 4-5 |
36 rows × 2 columns
Note
pd.read_csv uses the 'utf-8' encoding by default. All sample CSV files in this book are in the UTF-8 encoding, so there is no need to specify the encoding when reading them with pd.read_csv. Otherwise, the right encoding can be specified with the encoding parameter of pd.read_csv.
The next file we look into is 'routes.txt'. This file describes the public transport routes, identified by 'route_id' and also specifying the name of the route:
'route_short_name'—typically a number, such as bus line number'route_long_name'—the route textual description
It also contains 'agency_id' to identify the agency which operates each route:
cols = ['route_id', 'agency_id', 'route_short_name', 'route_long_name']
pd.read_csv('data/gtfs/routes.txt', usecols=cols)
| route_id | agency_id | route_short_name | route_long_name | |
|---|---|---|---|---|
| 0 | 1 | 25 | 1 | ת. רכבת יבנה מערב-יבנה<->ת. רכב... |
| 1 | 2 | 25 | 1 | ת. רכבת יבנה מזרח-יבנה<->ת. רכב... |
| 2 | 3 | 25 | 2 | ת. רכבת יבנה מערב-יבנה<->ת. רכב... |
| 3 | 4 | 25 | 2א | ת. רכבת יבנה מערב-יבנה<->ת. רכב... |
| 4 | 5 | 25 | 2 | ת. רכבת יבנה מזרח-יבנה<->ת. רכב... |
| ... | ... | ... | ... | ... |
| 8183 | 37315 | 2 | NaN | תא אוניברסיטה-תל אביב יפו<->באר... |
| 8184 | 37316 | 2 | NaN | תא אוניברסיטה-תל אביב יפו<->הרצ... |
| 8185 | 37342 | 6 | 46 | גן טכנולוגי-נצרת<->מסרארה חט''ב... |
| 8186 | 37343 | 6 | 46 | גן טכנולוגי-נצרת<->מסרארה חט''ב... |
| 8187 | 37350 | 2 | NaN | באר שבע מרכז-באר שבע<->נהריה-נהריה |
8188 rows × 4 columns
The 'routes.txt' table is linked to the 'trips.txt' table, which characterizes trips taking place at specific times of day for a given route. Trips are uniquely identified by 'trip_id' and linked with the routes table via the 'route_id' column. As you can see from the small subset below, each route may be associated with multiple trips. The trips table also has a 'shape_id' column which associates a trip with the geographical line geometry (see 'shapes.txt' below). You can see that trips of the same route at different times of day typically follow exactly the same geographical shape, thus having identical 'shape_id' values, but this is not guaranteed:
cols = ['route_id', 'trip_id', 'shape_id']
pd.read_csv('data/gtfs/trips.txt', usecols=cols)
| route_id | trip_id | shape_id | |
|---|---|---|---|
| 0 | 60 | 3764_081023 | 126055.0 |
| 1 | 68 | 3143332_011023 | 128020.0 |
| 2 | 68 | 3143338_011023 | 128020.0 |
| 3 | 68 | 3244955_011023 | 128020.0 |
| 4 | 68 | 3244956_011023 | 128020.0 |
| ... | ... | ... | ... |
| 398086 | 43 | 2827_081023 | 127560.0 |
| 398087 | 43 | 17254928_011023 | 127560.0 |
| 398088 | 43 | 2796_011023 | 127560.0 |
| 398089 | 43 | 2806_011023 | 127560.0 |
| 398090 | 43 | 2807_011023 | 127560.0 |
398091 rows × 3 columns
The 'stop_times.txt' table lists the stops associated with each trip (via the 'trip_id' column), including the (scheduled) time of arrival at each stop. The stop times table also includes the 'stop_id' column to associate it with the stop characteristics (see 'stops.txt' below). In addition, the 'shape_dist_traveled' column contains cumulative distance traveled (in \(m\)) at each stop. You can see the increasing distance traveled from one from stop to another (in the same trip) in the printout below. The 'stop_times.txt' table is the largest one, in terms of file size, among the GTFS files; keep in mind that it lists all stops, at all times of day, of all public transport routes:
cols = ['trip_id', 'arrival_time', 'departure_time', 'stop_id', 'shape_dist_traveled']
pd.read_csv('data/gtfs/stop_times.txt', usecols=cols)
| trip_id | arrival_time | departure_time | stop_id | shape_dist_traveled | |
|---|---|---|---|---|---|
| 0 | 1_011023 | 05:10:00 | 05:10:00 | 38725 | 0.0 |
| 1 | 1_011023 | 05:12:23 | 05:12:23 | 15582 | 714.0 |
| 2 | 1_011023 | 05:13:20 | 05:13:20 | 15583 | 1215.0 |
| 3 | 1_011023 | 05:14:31 | 05:14:31 | 16086 | 1791.0 |
| 4 | 1_011023 | 05:15:09 | 05:15:09 | 16085 | 2152.0 |
| ... | ... | ... | ... | ... | ... |
| 15169524 | 9999194_151023 | 20:19:45 | 20:19:45 | 9272 | 12666.0 |
| 15169525 | 9999194_151023 | 20:20:59 | 20:20:59 | 11495 | 12970.0 |
| 15169526 | 9999194_151023 | 20:21:59 | 20:21:59 | 9550 | 13237.0 |
| 15169527 | 9999194_151023 | 20:22:40 | 20:22:40 | 10483 | 13395.0 |
| 15169528 | 9999194_151023 | 20:23:08 | 20:23:08 | 11809 | 13597.0 |
15169529 rows × 5 columns
The 'stops.txt' table, which we already worked with earlier (see Reading CSV files), describes the fixed, i.e., unrelated to time of day, properties of the stops. It is linked with the 'stops_times.txt' table via the 'stop_id' column. Stop properties include the stop name ('stop_name'), and its coordinates ('stop_lon' and 'stop_lat'):
cols = ['stop_id', 'stop_name', 'stop_lat', 'stop_lon']
pd.read_csv('data/gtfs/stops.txt', usecols=cols)
| stop_id | stop_name | stop_lat | stop_lon | |
|---|---|---|---|---|
| 0 | 1 | בי''ס בר לב/בן יהודה | 32.183985 | 34.917554 |
| 1 | 2 | הרצל/צומת בילו | 31.870034 | 34.819541 |
| 2 | 3 | הנחשול/הדייגים | 31.984553 | 34.782828 |
| 3 | 4 | משה פריד/יצחק משקה | 31.888325 | 34.790700 |
| 4 | 6 | ת. מרכזית לוד/הורדה | 31.956392 | 34.898098 |
| ... | ... | ... | ... | ... |
| 34002 | 50182 | מסוף שכונת החותרים/הורדה | 32.748564 | 34.966691 |
| 34003 | 50183 | מסוף שכונת החותרים/איסוף | 32.748776 | 34.966844 |
| 34004 | 50184 | מדבריום | 31.260857 | 34.744373 |
| 34005 | 50185 | כביש 25/מדבריום | 31.263478 | 34.747607 |
| 34006 | 50186 | כביש 25/מדבריום | 31.264262 | 34.746768 |
34007 rows × 4 columns
Finally, the 'shapes.txt' table contains the trip geometries. It is linked with the 'trips.txt' table via the common 'shape_id' column.
cols = ['shape_id', 'shape_pt_lat', 'shape_pt_lon', 'shape_pt_sequence']
pd.read_csv('data/gtfs/shapes.txt', usecols=cols)
| shape_id | shape_pt_lat | shape_pt_lon | shape_pt_sequence | |
|---|---|---|---|---|
| 0 | 69895 | 32.164723 | 34.848813 | 1 |
| 1 | 69895 | 32.164738 | 34.848972 | 2 |
| 2 | 69895 | 32.164771 | 34.849177 | 3 |
| 3 | 69895 | 32.164841 | 34.849429 | 4 |
| 4 | 69895 | 32.164889 | 34.849626 | 5 |
| ... | ... | ... | ... | ... |
| 7546509 | 146260 | 32.084831 | 34.796385 | 953 |
| 7546510 | 146260 | 32.084884 | 34.796040 | 954 |
| 7546511 | 146260 | 32.084908 | 34.795895 | 955 |
| 7546512 | 146260 | 32.084039 | 34.795698 | 956 |
| 7546513 | 146260 | 32.083737 | 34.795645 | 957 |
7546514 rows × 4 columns
The relations between the six above-mentioned files in the GTFS dataset are summarized in Fig. 29.
Fig. 29 Relation between the six GTFS files listed in Table 15#
GTFS example#
Now that we have an idea about the structure and purpose of the GTFS dataset, let’s demonstrate join operations between its tables. This is a classical example where joining tables is essential: you cannot go very far with GTFS data analysis, or processing, without joining the various tables together (see Fig. 29).
To make things simple, in this example, consider just two of the tables in the GTFS dataset:
'agency.txt', where rows represent public transport operators'routes.txt', where rows represent public transport routes
As shown above (see What is GTFS?), the two tables share a common column 'agency_id', which we can use to join the two, to find out which public routes belong to each operator.
First, let’s import the 'agency.txt' table and subset the columns of interest, 'agency_id' and 'agency_name':
cols = ['agency_id', 'agency_name']
agency = pd.read_csv('data/gtfs/agency.txt', usecols=cols)
agency
| agency_id | agency_name | |
|---|---|---|
| 0 | 2 | רכבת ישראל |
| 1 | 3 | אגד |
| 2 | 4 | אלקטרה אפיקים תחבורה |
| 3 | 5 | דן |
| 4 | 6 | ש.א.מ |
| ... | ... | ... |
| 31 | 51 | ירושלים-צור באהר איחוד |
| 32 | 91 | מוניות מטרו קו |
| 33 | 93 | מוניות מאיה יצחק שדה |
| 34 | 97 | אודליה מוניות בעמ |
| 35 | 98 | מוניות רב קווית 4-5 |
36 rows × 2 columns
Second, we import the 'routes.txt' table, this time selecting the 'route_id', 'agency_id', 'route_short_name', and 'route_long_name' columns. Note the common 'agency_id' column, which we are going to use when joining the two tables:
cols = ['route_id', 'agency_id', 'route_short_name', 'route_long_name']
routes = pd.read_csv('data/gtfs/routes.txt', usecols=cols)
routes
| route_id | agency_id | route_short_name | route_long_name | |
|---|---|---|---|---|
| 0 | 1 | 25 | 1 | ת. רכבת יבנה מערב-יבנה<->ת. רכב... |
| 1 | 2 | 25 | 1 | ת. רכבת יבנה מזרח-יבנה<->ת. רכב... |
| 2 | 3 | 25 | 2 | ת. רכבת יבנה מערב-יבנה<->ת. רכב... |
| 3 | 4 | 25 | 2א | ת. רכבת יבנה מערב-יבנה<->ת. רכב... |
| 4 | 5 | 25 | 2 | ת. רכבת יבנה מזרח-יבנה<->ת. רכב... |
| ... | ... | ... | ... | ... |
| 8183 | 37315 | 2 | NaN | תא אוניברסיטה-תל אביב יפו<->באר... |
| 8184 | 37316 | 2 | NaN | תא אוניברסיטה-תל אביב יפו<->הרצ... |
| 8185 | 37342 | 6 | 46 | גן טכנולוגי-נצרת<->מסרארה חט''ב... |
| 8186 | 37343 | 6 | 46 | גן טכנולוגי-נצרת<->מסרארה חט''ב... |
| 8187 | 37350 | 2 | NaN | באר שבע מרכז-באר שבע<->נהריה-נהריה |
8188 rows × 4 columns
Now, we can join the routes and agency tables based on the common 'agency_id' column:
routes = pd.merge(routes, agency, on='agency_id', how='left')
routes
| route_id | agency_id | route_short_name | route_long_name | agency_name | |
|---|---|---|---|---|---|
| 0 | 1 | 25 | 1 | ת. רכבת יבנה מערב-יבנה<->ת. רכב... | אלקטרה אפיקים |
| 1 | 2 | 25 | 1 | ת. רכבת יבנה מזרח-יבנה<->ת. רכב... | אלקטרה אפיקים |
| 2 | 3 | 25 | 2 | ת. רכבת יבנה מערב-יבנה<->ת. רכב... | אלקטרה אפיקים |
| 3 | 4 | 25 | 2א | ת. רכבת יבנה מערב-יבנה<->ת. רכב... | אלקטרה אפיקים |
| 4 | 5 | 25 | 2 | ת. רכבת יבנה מזרח-יבנה<->ת. רכב... | אלקטרה אפיקים |
| ... | ... | ... | ... | ... | ... |
| 8183 | 37315 | 2 | NaN | תא אוניברסיטה-תל אביב יפו<->באר... | רכבת ישראל |
| 8184 | 37316 | 2 | NaN | תא אוניברסיטה-תל אביב יפו<->הרצ... | רכבת ישראל |
| 8185 | 37342 | 6 | 46 | גן טכנולוגי-נצרת<->מסרארה חט''ב... | ש.א.מ |
| 8186 | 37343 | 6 | 46 | גן טכנולוגי-נצרת<->מסרארה חט''ב... | ש.א.מ |
| 8187 | 37350 | 2 | NaN | באר שבע מרכז-באר שבע<->נהריה-נהריה | רכבת ישראל |
8188 rows × 5 columns
Now we have the additional 'agency_name' column in the routes table, which tells us the name of the operator for each route.
Note
For more information and examples of pd.merge, check out the following official tutorials from the pandas documentation:
Aggregation#
Overview#
Aggregation is commonly used to summarize tabular data. In an aggregation procedure, a function is applied on subsets of table rows, whereas subsets are specified through one or more grouping variables, to obtain a new (smaller) table with summaries per group. Aggregation is one of the techniques following the split-apply-combine concept.
In pandas, there are several different ways of specifying the function(s) to be applied on the table column(s). We will go over three commonly used scenarios:
Afterwards, we are going to introduce a useful shortcut for a particularly common type of aggregation: counting the occurence of each unique value in a column (see Value counts).
Same function on all columns#
For a minimal example of aggregation, let’s go back to the small stations table:
stations
| name | city | lines | piano | lon | lat | |
|---|---|---|---|---|---|---|
| 0 | Beer-Sheva Center | Beer-Sheva | 4 | False | 34.798443 | 31.243288 |
| 1 | Beer-Sheva University | Beer-Sheva | 5 | True | 34.812831 | 31.260284 |
| 2 | Dimona | Dimona | 1 | False | 35.011635 | 31.068616 |
Suppose that we are interested in the number of lines (the lines column) and the number of stations with a piano (the piano column), which involves summing the values in those two columns. We already know how the totals, i.e., sums of each column, can be calculated (see Row/col-wise operations):
stations[['lines', 'piano']].sum()
lines 10
piano 1
dtype: int64
However, what if we wanted to calculate the number of lines and stations with a piano per town, separately? To do that, we basically need to split the table according to the 'city' column, then apply the .sum method on each subset. Here is how this can be done. Instead of simply using .sum, we split (or “group”) the table by 'city', and only then apply the .sum method. Splitting is done using the .groupby method, which accepts the column name (or list of column names) that specifies the grouping:
stations.groupby('city')[['lines', 'piano']].sum()
| lines | piano | |
|---|---|---|
| city | ||
| Beer-Sheva | 9 | 1 |
| Dimona | 1 | 0 |
Note that the grouping variable(s) become the index of the resulting DataFrame. For our purposes, we are usually going to prefer to work with the grouping variable in its own column. In such case, we end the expression with .reset_index() (see Resetting the index):
stations.groupby('city')[['lines', 'piano']].sum().reset_index()
| city | lines | piano | |
|---|---|---|---|
| 0 | Beer-Sheva | 9 | 1 |
| 1 | Dimona | 1 | 0 |
Note
Tables can also be grouped by two or more columns, in which case the column names are passed in a list, as in dat.groupby(['a','b']), where 'a' and 'b' are column names in dat.
The .sum method is just one example, there are many other useful functions we can use, such as the ones listed in Table 14. Additional methods specific to .groupby are .first and .last, to select the first and last row per group. For example, to get the maximum per group we can combine .groupby with .max:
stations.groupby('city').max().reset_index()
| city | name | lines | piano | lon | lat | |
|---|---|---|---|---|---|---|
| 0 | Beer-Sheva | Beer-Sheva University | 5 | True | 34.812831 | 31.260284 |
| 1 | Dimona | Dimona | 1 | False | 35.011635 | 31.068616 |
For a more realistic example, consider the routes table:
routes
| route_id | agency_id | route_short_name | route_long_name | agency_name | |
|---|---|---|---|---|---|
| 0 | 1 | 25 | 1 | ת. רכבת יבנה מערב-יבנה<->ת. רכב... | אלקטרה אפיקים |
| 1 | 2 | 25 | 1 | ת. רכבת יבנה מזרח-יבנה<->ת. רכב... | אלקטרה אפיקים |
| 2 | 3 | 25 | 2 | ת. רכבת יבנה מערב-יבנה<->ת. רכב... | אלקטרה אפיקים |
| 3 | 4 | 25 | 2א | ת. רכבת יבנה מערב-יבנה<->ת. רכב... | אלקטרה אפיקים |
| 4 | 5 | 25 | 2 | ת. רכבת יבנה מזרח-יבנה<->ת. רכב... | אלקטרה אפיקים |
| ... | ... | ... | ... | ... | ... |
| 8183 | 37315 | 2 | NaN | תא אוניברסיטה-תל אביב יפו<->באר... | רכבת ישראל |
| 8184 | 37316 | 2 | NaN | תא אוניברסיטה-תל אביב יפו<->הרצ... | רכבת ישראל |
| 8185 | 37342 | 6 | 46 | גן טכנולוגי-נצרת<->מסרארה חט''ב... | ש.א.מ |
| 8186 | 37343 | 6 | 46 | גן טכנולוגי-נצרת<->מסרארה חט''ב... | ש.א.מ |
| 8187 | 37350 | 2 | NaN | באר שבע מרכז-באר שבע<->נהריה-נהריה | רכבת ישראל |
8188 rows × 5 columns
We can use aggregation with the .nunique function (Table 14) to calculate the number of unique route IDs that each public transport agency operates, as follows:
tmp = routes.groupby('agency_name')[['route_id']].nunique().reset_index()
tmp
| agency_name | route_id | |
|---|---|---|
| 0 | אגד | 1621 |
| 1 | אודליה מוניות בעמ | 2 |
| 2 | אלקטרה אפיקים | 413 |
| 3 | אלקטרה אפיקים תחבורה | 297 |
| 4 | אקסטרה | 65 |
| ... | ... | ... |
| 31 | קווים | 1161 |
| 32 | רכבת ישראל | 1008 |
| 33 | ש.א.מ | 158 |
| 34 | תבל | 5 |
| 35 | תנופה | 287 |
36 rows × 2 columns
To find out which agencies have the most routes, the resulting table can be sorted using .sort_values combined with ascending=False (see Sorting):
tmp.sort_values('route_id', ascending=False)
| agency_name | route_id | |
|---|---|---|
| 0 | אגד | 1621 |
| 31 | קווים | 1161 |
| 32 | רכבת ישראל | 1008 |
| 27 | מטרופולין | 691 |
| 29 | נתיב אקספרס | 577 |
| ... | ... | ... |
| 20 | כפיר | 3 |
| 1 | אודליה מוניות בעמ | 2 |
| 19 | כבל אקספרס | 2 |
| 21 | כרמלית | 2 |
| 22 | מוניות מאיה יצחק שדה | 1 |
36 rows × 2 columns
Exercise 06-c
Calculate a series of global temperature means for each decade, based on the information in
ZonAnn.Ts+dSST.csv.First, you need to create a new variable named
decadebased on the'Year', using an expression such as the one shown below.Second, use
.groupbyto calculate the mean global temperature in each decade.
dat['Year'] // 10 * 10
Exercise 06-d
The
'routes.txt'table describes public transport routes (uniquely identified byroute_id). The'trips.txt'table, however, descibes specific journeys of a along those routes, at a specific time of day (uniquely identified bytrip_id). The two tables are associated with one another via the commonroute_idcolumn (see What is GTFS?). For example, a route that operates five times a day will be represented in one row in the'routes.txt'table, and five rows in the'trips.txt'table.Join
'routes.txt'to'trips.txt', using the commonroute_idcolumn.Calculate a table with the number of unique
trip_id’s perroute_id.Sort the table according to number of unique trips, and print a summary with the numbers and names (
route_short_nameandroute_long_namecolumns fromroutes.txt, respectively) of the most and least frequent routes (Fig. 30).
| route_id | route_short_name | route_long_name | trip_id | |
|---|---|---|---|---|
| 2837 | 11685 | 1 | ת. מרכזית חוף הכרמל/רציפים עירו... | 1344 |
| 2839 | 11689 | 1 | מרכזית הקריות-קרית מוצקין<->ת. ... | 1300 |
| 6465 | 29603 | 10 | אוניברסיטת חיפה-חיפה<->מרכזית ה... | 1201 |
| 6464 | 29602 | 10 | מרכזית המפרץ-חיפה<->אוניברסיטת ... | 1201 |
| 3192 | 12405 | 15 | תחנה תפעולית/ביטוח לאומי-ירושלי... | 978 |
| ... | ... | ... | ... | ... |
| 3227 | 12643 | 294 | מסעף מעלות צפון-מעלות תרשיחא<->... | 1 |
| 122 | 344 | 39 | עירייה-מעלות תרשיחא<->ת. מרכזית... | 1 |
| 153 | 412 | 82 | הרבי מלובאוויטש/כ''ב ילדי מעלות... | 1 |
| 114 | 320 | 25 | גורן 4-גורן<->ת. מרכזית נהריה/ה... | 1 |
| 4136 | 16410 | 300 | כביש 436/גבעת זאב-גבעת זאב<->שד... | 1 |
7180 rows × 4 columns
Fig. 30 Solution of exercise-06-c#
Different functions (agg)#
Sometimes we need to aggregate a DataFrame while applying a different function on each column, rather than the same function on all columns. For the next example, consider the 'world_cities.csv' file, which has information about major cities in the world:
cities = pd.read_csv('data/world_cities.csv')
cities
| city | country | pop | lat | lon | capital | |
|---|---|---|---|---|---|---|
| 0 | 'Abasan al-Jadidah | Palestine | 5629 | 31.31 | 34.34 | 0 |
| 1 | 'Abasan al-Kabirah | Palestine | 18999 | 31.32 | 34.35 | 0 |
| 2 | 'Abdul Hakim | Pakistan | 47788 | 30.55 | 72.11 | 0 |
| 3 | 'Abdullah-as-Salam | Kuwait | 21817 | 29.36 | 47.98 | 0 |
| 4 | 'Abud | Palestine | 2456 | 32.03 | 35.07 | 0 |
| ... | ... | ... | ... | ... | ... | ... |
| 43640 | az-Zubayr | Iraq | 124611 | 30.39 | 47.71 | 0 |
| 43641 | az-Zulfi | Saudi Arabia | 54070 | 26.30 | 44.80 | 0 |
| 43642 | az-Zuwaytinah | Libya | 21984 | 30.95 | 20.12 | 0 |
| 43643 | s-Gravenhage | Netherlands | 479525 | 52.07 | 4.30 | 0 |
| 43644 | s-Hertogenbosch | Netherlands | 135529 | 51.68 | 5.30 | 0 |
43645 rows × 6 columns
Suppose we want to summarize the information per country ('country' column). However, we want each column to be treated differently:
The city names (
'city'column) will be countedPopulation (
'pop'column) will be summedLongitude and latitude (
'lon'and'lat'columns, respectively) will be averaged
This can be done using the .agg method applied on a grouped DataFrame instead of a specific method (such as .nunique). The .agg method accepts a dictionary, with key:value pairs of the form 'column':function, where:
'column'—Name of the column to be aggregatedfunction—The function to apply on each group in that column. This can be:a string referring to a
pandasmethod (such as'nunique', see Table 14)a standalone function that can be applied on a
Series, such asnp.meanornp.sum
In our case, the aggregation expression is as follows. As a result, we get a new table where each row represents a country, with the count of unique city names, sum of the population across all cities, and average longitute and latitude across all cities:
cities.groupby('country').agg({
'city': 'nunique',
'pop': 'sum',
'lon': 'mean',
'lat': 'mean'
}).reset_index()
| country | city | pop | lon | lat | |
|---|---|---|---|---|---|
| 0 | Afghanistan | 115 | 7543856 | 66.920342 | 34.807692 |
| 1 | Albania | 67 | 1536232 | 19.975522 | 41.087463 |
| 2 | Algeria | 315 | 20508642 | 3.535443 | 35.467025 |
| 3 | American Samoa | 35 | 58021 | -170.663714 | -14.299143 |
| 4 | Andorra | 7 | 69031 | 1.538571 | 42.534286 |
| ... | ... | ... | ... | ... | ... |
| 234 | Wallis and Futuna | 23 | 11380 | -176.585652 | -13.512609 |
| 235 | Western Sahara | 4 | 338786 | -13.820000 | 25.935000 |
| 236 | Yemen | 30 | 5492077 | 45.088667 | 14.481000 |
| 237 | Zambia | 73 | 4032170 | 28.179315 | -13.404247 |
| 238 | Zimbabwe | 76 | 4231859 | 30.442500 | -18.629211 |
239 rows × 5 columns
Custom functions (agg)#
Finally, sometimes we need to apply a custom function when aggregating a table. For example, suppose that we need to calculate the population difference between the most populated, and least populated, cities, in each country. To do that, we can define our own function, named diff_max_min, which:
accepts a
Seriesnamedx, andreturns the difference between the maximum and minimum,
x.max()-x.min().
Here is the function definition:
def diff_max_min(x):
return x.max() - x.min()
It is worthwhile to test the function, to make sure it works as expected:
diff_max_min(pd.Series([15, 20, 4, 12, 6]))
np.int64(16)
Now, we can use the function inside the .agg expression, just like any other predefined function:
cities.groupby('country').agg({
'city': 'nunique',
'pop': diff_max_min
}).reset_index()
| country | city | pop | |
|---|---|---|---|
| 0 | Afghanistan | 115 | 3117579 |
| 1 | Albania | 67 | 379803 |
| 2 | Algeria | 315 | 2024344 |
| 3 | American Samoa | 35 | 11341 |
| 4 | Andorra | 7 | 17740 |
| ... | ... | ... | ... |
| 234 | Wallis and Futuna | 23 | 1142 |
| 235 | Western Sahara | 4 | 146906 |
| 236 | Yemen | 30 | 1918607 |
| 237 | Zambia | 73 | 1305210 |
| 238 | Zimbabwe | 76 | 1574930 |
239 rows × 3 columns
For example, here is how the “new” pop value is obtained for the first country ('Afghanistan'):
diff_max_min(cities.loc[cities['country'] == 'Afghanistan', 'pop'])
np.int64(3117579)
which is equivalent to:
pop = cities.loc[cities['country'] == 'Afghanistan', 'pop']
pop.max() - pop.min()
np.int64(3117579)
Alternatively, we can use a lambda function (see Lambda functions), to do the same, using more concise syntax:
cities.groupby('country').agg({
'city': 'nunique',
'pop': lambda x: x.max() - x.min()
}).reset_index()
| country | city | pop | |
|---|---|---|---|
| 0 | Afghanistan | 115 | 3117579 |
| 1 | Albania | 67 | 379803 |
| 2 | Algeria | 315 | 2024344 |
| 3 | American Samoa | 35 | 11341 |
| 4 | Andorra | 7 | 17740 |
| ... | ... | ... | ... |
| 234 | Wallis and Futuna | 23 | 1142 |
| 235 | Western Sahara | 4 | 146906 |
| 236 | Yemen | 30 | 1918607 |
| 237 | Zambia | 73 | 1305210 |
| 238 | Zimbabwe | 76 | 1574930 |
239 rows × 3 columns
Note
For more information and examples on aggregation, and other types of split-apply-combine operations which we have not mentioned, see https://pandas.pydata.org/docs/user_guide/groupby.html#aggregation.
Value counts#
A very common type of aggregation is to calculate how many times each unique value appears in a Series. For example, recall the way that we calculated the number of unique routes per agency:
routes \
.groupby('agency_name')[['route_id']] \
.nunique() \
.reset_index() \
.sort_values('route_id', ascending=False)
| agency_name | route_id | |
|---|---|---|
| 0 | אגד | 1621 |
| 31 | קווים | 1161 |
| 32 | רכבת ישראל | 1008 |
| 27 | מטרופולין | 691 |
| 29 | נתיב אקספרס | 577 |
| ... | ... | ... |
| 20 | כפיר | 3 |
| 1 | אודליה מוניות בעמ | 2 |
| 19 | כבל אקספרס | 2 |
| 21 | כרמלית | 2 |
| 22 | מוניות מאיה יצחק שדה | 1 |
36 rows × 2 columns
The .value_counts method which can be thought of as a shortcut (assuming that all route_id values are unique, which is true in this case):
routes[['agency_name']].value_counts()
agency_name
אגד 1621
קווים 1161
רכבת ישראל 1008
מטרופולין 691
נתיב אקספרס 577
...
כפיר 3
אודליה מוניות בעמ 2
כבל אקספרס 2
כרמלית 2
מוניות מאיה יצחק שדה 1
Name: count, Length: 36, dtype: int64
The result is a Series where the index is the agency name, and the values are the counts. It tells us how many rows are there in the routes table, for each agency. For example, 'אגד' is repeated over 1621 rows.
As you can see, the resuling Series is sorted by count, from highest to lowest. In case we need to sort by index, i.e., by agency name, we can use the .sort_index method:
routes[['agency_name']].value_counts().sort_index()
agency_name
אגד 1621
אודליה מוניות בעמ 2
אלקטרה אפיקים 413
אלקטרה אפיקים תחבורה 297
אקסטרה 65
...
קווים 1161
רכבת ישראל 1008
ש.א.מ 158
תבל 5
תנופה 287
Name: count, Length: 36, dtype: int64
Exercise 06-e
How often is there more than one shape (
shape_id) for the same route (route_id)?Use the
'trips.txt'table to calculate the number of unique values ofshape_idfor eachroute_id.Summarize the resulting column to calculate how many
route_ids has0,1,2, etc. ofshape_id.
More exercises#
Exercise 06-f
Read the
'agency.txt','routes.txt','trips.txt', and'shapes.txt'tables (located in thegtfsdirectory).Subset just the
'דן באר שבע'operator routes from theroutestable (first find out itsagency_idin theagencytable)Subset only those
routeswhere'route_short_name'is'24'(i.e., routes of bus'24')Join the routes table with
tripsand filter to retain just one trip (the firsttrip_id)Join the trip table with
shapesPrint the resulting table with the shape coordinates of one of the trips of bus
'24'(Fig. 31)Create a scatterplot of the trip coordinates, with
'shape_pt_lon'values on the x-axis and'shape_pt_lat'values on the y-axis (Fig. 32)
| route_id | route_short_name | route_long_name | trip_id | shape_id | |
|---|---|---|---|---|---|
| 0 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 26385618_011023 | 136236.0 |
| 1 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 26385620_011023 | 136236.0 |
| 2 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 25887153_011023 | 136236.0 |
| 3 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 29025527_011023 | 136236.0 |
| 4 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 25887168_011023 | 136236.0 |
| ... | ... | ... | ... | ... | ... |
| 142 | 17538 | 24 | מסוף רמות-באר שבע<->ת.מרכזית/עי... | 22834961_011023 | 127348.0 |
| 143 | 17538 | 24 | מסוף רמות-באר שבע<->ת.מרכזית/עי... | 22834957_011023 | 127348.0 |
| 144 | 17538 | 24 | מסוף רמות-באר שבע<->ת.מרכזית/עי... | 26385928_011023 | 127348.0 |
| 145 | 17538 | 24 | מסוף רמות-באר שבע<->ת.מרכזית/עי... | 26385947_011023 | 127348.0 |
| 146 | 17538 | 24 | מסוף רמות-באר שבע<->ת.מרכזית/עי... | 26385948_011023 | 127348.0 |
147 rows × 5 columns
Fig. 31 Solution of exercise-06-f: Coordinates of the 1st trip of bus '24' by 'דן באר שבע'#
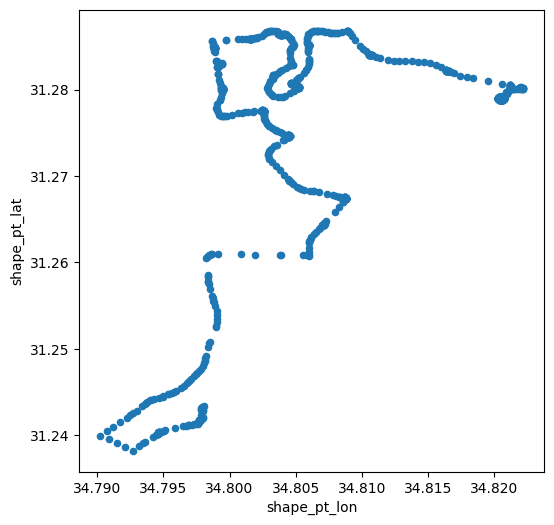
Fig. 32 Solution of exercise-06-f: Coordinates of the 1st trip of bus '24' by 'דן באר שבע'#
Exercise 06-g
In this exercise, you are going to find the names of the five longest bus routes (in terms of distance traveled) in Israel. To do that, go through the following steps.
Read the
stop_times.txttable and select just the'trip_id'and'shape_dist_traveled'columns.Calculate the maximum (i.e., total) distance traveled (
'shape_dist_traveled') for each public transit trip ('trip_id') (Fig. 33).Read the
trips.txttable and select just the'trip_id', and'route_id'.Join the resulting
tripstable with thestop_timestable from the previous step (using the common"trip_id"column), to get the'route_id'value of each trip.Retain just one (the first) trip per
'route_id'(Fig. 34).Read the
routes.txttable and subset the'route_id','route_short_name', and'route_long_name'columns.Join the resulting table with the trips table (using the common
'route_id'column), to get the'route_short_name'and'route_long_name'values of each route (Fig. 35).Sort the table according to
'shape_dist_traveled'(from largest to smallest), and print the first five rows (Fig. 36). Those are the longest-distance public transport routes in the GTFS dataset.Convert the
'route_long_name'values of the first five rows from the previous step, collect them into alist, and print thelist, to see the full names of the routes. You can see that all five routes are from Haifa to Eilat or the other way around.
| trip_id | shape_dist_traveled | |
|---|---|---|
| 0 | 10004214_011023 | 11152.0 |
| 1 | 10004215_011023 | 11152.0 |
| 2 | 1001407_011023 | 15061.0 |
| 3 | 10015281_071023 | 21688.0 |
| 4 | 10015281_081023 | 21688.0 |
| ... | ... | ... |
| 398086 | 9998411_091023 | 14147.0 |
| 398087 | 9998411_131023 | 14040.0 |
| 398088 | 9999194_061023 | 13597.0 |
| 398089 | 9999194_081023 | 13597.0 |
| 398090 | 9999194_151023 | 13597.0 |
398091 rows × 2 columns
Fig. 33 Solution of exercise-06-g: Total distance traveled in each trip, based on stop_times.txt#
| route_id | trip_id | shape_dist_traveled | |
|---|---|---|---|
| 0 | 1 | 17336167_081023 | 7258.0 |
| 1 | 2 | 13076786_081023 | 7088.0 |
| 2 | 3 | 5655904_011023 | 9883.0 |
| 3 | 4 | 585026983_011023 | 7025.0 |
| 4 | 5 | 5656121_081023 | 9750.0 |
| ... | ... | ... | ... |
| 8183 | 37315 | 1_88437 | NaN |
| 8184 | 37316 | 1_88438 | NaN |
| 8185 | 37342 | 585305994_121023 | 16432.0 |
| 8186 | 37343 | 585305995_121023 | 16761.0 |
| 8187 | 37350 | 1_88086 | NaN |
8188 rows × 3 columns
Fig. 34 Solution of exercise-06-g: Joined with trips.txt, selecting just the 1st trips per route_id#
| route_id | route_short_name | route_long_name | trip_id | shape_dist_traveled | |
|---|---|---|---|---|---|
| 0 | 1 | 1 | ת. רכבת יבנה מערב-יבנה<->ת. רכב... | 17336167_081023 | 7258.0 |
| 1 | 2 | 1 | ת. רכבת יבנה מזרח-יבנה<->ת. רכב... | 13076786_081023 | 7088.0 |
| 2 | 3 | 2 | ת. רכבת יבנה מערב-יבנה<->ת. רכב... | 5655904_011023 | 9883.0 |
| 3 | 4 | 2א | ת. רכבת יבנה מערב-יבנה<->ת. רכב... | 585026983_011023 | 7025.0 |
| 4 | 5 | 2 | ת. רכבת יבנה מזרח-יבנה<->ת. רכב... | 5656121_081023 | 9750.0 |
| ... | ... | ... | ... | ... | ... |
| 8183 | 37315 | NaN | תא אוניברסיטה-תל אביב יפו<->באר... | 1_88437 | NaN |
| 8184 | 37316 | NaN | תא אוניברסיטה-תל אביב יפו<->הרצ... | 1_88438 | NaN |
| 8185 | 37342 | 46 | גן טכנולוגי-נצרת<->מסרארה חט''ב... | 585305994_121023 | 16432.0 |
| 8186 | 37343 | 46 | גן טכנולוגי-נצרת<->מסרארה חט''ב... | 585305995_121023 | 16761.0 |
| 8187 | 37350 | NaN | באר שבע מרכז-באר שבע<->נהריה-נהריה | 1_88086 | NaN |
8188 rows × 5 columns
Fig. 35 Solution of exercise-06-g: Joined with routes.txt#
| route_id | route_short_name | route_long_name | trip_id | shape_dist_traveled | |
|---|---|---|---|---|---|
| 5155 | 19964 | 991 | ת. מרכזית חוף הכרמל/רציפים בינע... | 30526427_091023 | 446163.0 |
| 5475 | 21726 | 991 | ת. מרכזית אילת/רציפים-אילת<->ת.... | 30526417_061023 | 445475.0 |
| 1456 | 7307 | 993 | ת. מרכזית המפרץ/רציפים בינעירונ... | 2125973_151023 | 432200.0 |
| 1455 | 7305 | 993 | ת. מרכזית אילת/רציפים-אילת<->ת.... | 584673295_011023 | 432187.0 |
| 1153 | 5182 | 399 | ת. מרכזית חדרה/רציפים-חדרה<->ת.... | 584660100_091023 | 401875.0 |
Fig. 36 Solution of exercise-06-g: Sorted by shape_dist_traveled, from largest to smallest#
Exercise solutions#
Exercise 06-b#
import numpy as np
import pandas as pd
# Read data
dat = pd.read_csv('data/ZonAnn.Ts+dSST.csv')
cols = ['90S-64S', '64S-44S', '44S-24S', '24S-EQU', 'EQU-24N', '24N-44N', '44N-64N', '64N-90N']
regions = dat[cols]
# Function to calculate linear slope
def f(x, y):
return np.polyfit(x, y, 1)[0]
f(dat['Year'], regions['44S-24S'])
np.float64(0.006830158347399733)
# Calculate slopes per region
regions.apply(lambda i: f(dat['Year'], i), axis=0)
90S-64S 0.006262
64S-44S 0.005188
44S-24S 0.006830
24S-EQU 0.006690
EQU-24N 0.006562
24N-44N 0.008564
44N-64N 0.012204
64N-90N 0.018884
dtype: float64
Exercise 06-d#
import pandas as pd
# Read
routes = pd.read_csv('data/gtfs/routes.txt')
trips = pd.read_csv('data/gtfs/trips.txt')
# Join
dat = pd.merge(trips, routes, on='route_id', how='left')
# Calculate number of unique trips per route
dat = dat.groupby(['route_id', 'route_short_name', 'route_long_name']).nunique()['trip_id'].reset_index()
# Sort
dat = dat.sort_values('trip_id', ascending=False)
dat
Exercise 06-e#
import pandas as pd
trips = pd.read_csv('data/gtfs/trips.txt')
trips.groupby('route_id')['shape_id'].nunique().value_counts().sort_index()
Exercise 06-f#
import pandas as pd
# Read
agency = pd.read_csv('data/gtfs/agency.txt')
routes = pd.read_csv('data/gtfs/routes.txt')
trips = pd.read_csv('data/gtfs/trips.txt')
shapes = pd.read_csv('data/gtfs/shapes.txt')
# Subset
agency = agency[['agency_id', 'agency_name']]
routes = routes[['route_id', 'agency_id', 'route_short_name', 'route_long_name']]
trips = trips[['trip_id', 'route_id', 'shape_id']]
shapes = shapes[['shape_id', 'shape_pt_sequence', 'shape_pt_lon', 'shape_pt_lat']]
# Filter agency
agency = agency[agency['agency_name'] == 'דן באר שבע']
routes = routes[routes['agency_id'] == agency['agency_id'].iloc[0]]
routes = routes.drop('agency_id', axis=1)
# Filter route
routes = routes[routes['route_short_name'] == '24']
# Filter trip
routes = pd.merge(routes, trips, on='route_id', how='left')
trip = routes[routes['trip_id'] == routes['trip_id'].iloc[0]]
# Join with 'shapes'
trip = pd.merge(trip, shapes, on='shape_id', how='left')
trip = trip.sort_values(by='shape_pt_sequence')
trip
| route_id | route_short_name | route_long_name | trip_id | shape_id | shape_pt_sequence | shape_pt_lon | shape_pt_lat | |
|---|---|---|---|---|---|---|---|---|
| 0 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 26385618_011023 | 136236.0 | 1 | 34.797979 | 31.242009 |
| 1 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 26385618_011023 | 136236.0 | 2 | 34.797988 | 31.242103 |
| 2 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 26385618_011023 | 136236.0 | 3 | 34.797982 | 31.242829 |
| 3 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 26385618_011023 | 136236.0 | 4 | 34.797986 | 31.242980 |
| 4 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 26385618_011023 | 136236.0 | 5 | 34.798000 | 31.243121 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 856 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 26385618_011023 | 136236.0 | 857 | 34.822166 | 31.280183 |
| 857 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 26385618_011023 | 136236.0 | 858 | 34.822116 | 31.280225 |
| 858 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 26385618_011023 | 136236.0 | 859 | 34.822038 | 31.280246 |
| 859 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 26385618_011023 | 136236.0 | 860 | 34.821949 | 31.280250 |
| 860 | 17537 | 24 | ת.מרכזית/עירוניים לצפון-באר שבע... | 26385618_011023 | 136236.0 | 861 | 34.821860 | 31.280233 |
861 rows × 8 columns
trip.plot.scatter(x='shape_pt_lon', y='shape_pt_lat');

Exercise 06-g#
import pandas as pd
# Get total distance traveled per 'trip_id'
stop_times = pd.read_csv('data/gtfs/stop_times.txt')
stop_times = stop_times.groupby('trip_id')['shape_dist_traveled'].max()
stop_times = stop_times.reset_index()
stop_times
| trip_id | shape_dist_traveled | |
|---|---|---|
| 0 | 10004214_011023 | 11152.0 |
| 1 | 10004215_011023 | 11152.0 |
| 2 | 1001407_011023 | 15061.0 |
| 3 | 10015281_071023 | 21688.0 |
| 4 | 10015281_081023 | 21688.0 |
| ... | ... | ... |
| 398086 | 9998411_091023 | 14147.0 |
| 398087 | 9998411_131023 | 14040.0 |
| 398088 | 9999194_061023 | 13597.0 |
| 398089 | 9999194_081023 | 13597.0 |
| 398090 | 9999194_151023 | 13597.0 |
398091 rows × 2 columns
# Join with trips to get 'route_id' per trip
trips = pd.read_csv('data/gtfs/trips.txt')
trips = trips[['trip_id', 'route_id']]
trips = pd.merge(trips, stop_times, on='trip_id')
trips = trips.groupby('route_id').first()
trips = trips.reset_index()
trips
| route_id | trip_id | shape_dist_traveled | |
|---|---|---|---|
| 0 | 1 | 17336167_081023 | 7258.0 |
| 1 | 2 | 13076786_081023 | 7088.0 |
| 2 | 3 | 5655904_011023 | 9883.0 |
| 3 | 4 | 585026983_011023 | 7025.0 |
| 4 | 5 | 5656121_081023 | 9750.0 |
| ... | ... | ... | ... |
| 8183 | 37315 | 1_88437 | NaN |
| 8184 | 37316 | 1_88438 | NaN |
| 8185 | 37342 | 585305994_121023 | 16432.0 |
| 8186 | 37343 | 585305995_121023 | 16761.0 |
| 8187 | 37350 | 1_88086 | NaN |
8188 rows × 3 columns
# Join with routes to get route short/long names
routes = pd.read_csv('data/gtfs/routes.txt')
routes = routes[['route_id', 'route_short_name', 'route_long_name']]
routes = pd.merge(routes, trips, on='route_id')
routes
| route_id | route_short_name | route_long_name | trip_id | shape_dist_traveled | |
|---|---|---|---|---|---|
| 0 | 1 | 1 | ת. רכבת יבנה מערב-יבנה<->ת. רכב... | 17336167_081023 | 7258.0 |
| 1 | 2 | 1 | ת. רכבת יבנה מזרח-יבנה<->ת. רכב... | 13076786_081023 | 7088.0 |
| 2 | 3 | 2 | ת. רכבת יבנה מערב-יבנה<->ת. רכב... | 5655904_011023 | 9883.0 |
| 3 | 4 | 2א | ת. רכבת יבנה מערב-יבנה<->ת. רכב... | 585026983_011023 | 7025.0 |
| 4 | 5 | 2 | ת. רכבת יבנה מזרח-יבנה<->ת. רכב... | 5656121_081023 | 9750.0 |
| ... | ... | ... | ... | ... | ... |
| 8183 | 37315 | NaN | תא אוניברסיטה-תל אביב יפו<->באר... | 1_88437 | NaN |
| 8184 | 37316 | NaN | תא אוניברסיטה-תל אביב יפו<->הרצ... | 1_88438 | NaN |
| 8185 | 37342 | 46 | גן טכנולוגי-נצרת<->מסרארה חט''ב... | 585305994_121023 | 16432.0 |
| 8186 | 37343 | 46 | גן טכנולוגי-נצרת<->מסרארה חט''ב... | 585305995_121023 | 16761.0 |
| 8187 | 37350 | NaN | באר שבע מרכז-באר שבע<->נהריה-נהריה | 1_88086 | NaN |
8188 rows × 5 columns
# Sort accordinge to distance traveled
routes = routes.sort_values('shape_dist_traveled', ascending=False).head()
routes
| route_id | route_short_name | route_long_name | trip_id | shape_dist_traveled | |
|---|---|---|---|---|---|
| 5155 | 19964 | 991 | ת. מרכזית חוף הכרמל/רציפים בינע... | 30526427_091023 | 446163.0 |
| 5475 | 21726 | 991 | ת. מרכזית אילת/רציפים-אילת<->ת.... | 30526417_061023 | 445475.0 |
| 1456 | 7307 | 993 | ת. מרכזית המפרץ/רציפים בינעירונ... | 2125973_151023 | 432200.0 |
| 1455 | 7305 | 993 | ת. מרכזית אילת/רציפים-אילת<->ת.... | 584673295_011023 | 432187.0 |
| 1153 | 5182 | 399 | ת. מרכזית חדרה/רציפים-חדרה<->ת.... | 584660100_091023 | 401875.0 |
routes.head()["route_long_name"].to_list()
['ת. מרכזית חוף הכרמל/רציפים בינעירוני-חיפה<->ת. מרכזית אילת/הורדה-אילת-2#',
'ת. מרכזית אילת/רציפים-אילת<->ת. מרכזית חוף הכרמל/הורדה-חיפה-1#',
'ת. מרכזית המפרץ/רציפים בינעירוני-חיפה<->ת. מרכזית אילת/הורדה-אילת-2#',
'ת. מרכזית אילת/רציפים-אילת<->ת. מרכזית המפרץ/הורדה-חיפה-1#',
'ת. מרכזית חדרה/רציפים-חדרה<->ת. מרכזית אילת/הורדה-אילת-1#']