Chapter 5 Matrices and rasters
Last updated: 2020-08-12 00:35:51
Aims
Our aims in this chapter are:
- Start working with spatial data (rasters)
- Install and use packages beyond “base R”
- Introduce the basic
matrixandarraydata structures, and their analogous spatial data structure (classstars) for single band and multi-band rasters, respectively - Learn to access the cell values and other properties of rasters
- Learn to read and write raster data
We will use the following R packages:
starsmapviewcubeview
5.1 Matrices
5.1.1 What is a matrix?
A matrix is a two-dimensional collection of values of the same type (like a vector), where the number of values in all columns is equal. It is important to know how to work with matrices because it is a commonly used data structure, with many uses in data processing and analysis, including spatial data. For example, many R function accept a matrix as an argument, or return a matrix as a returned object, e.g., st_intersects (Section 8.3.3).
5.1.2 Creating a matrix
A matrix can be created with the matrix function. The matrix function accepts the following arguments:
data—A vector of the values to fill into the matrixnrow—The number of rowsncol—The number of columnsbyrow—Whether the matrix is filled by column (FALSE, the default) or by row (TRUE)
For example:
The nrow and ncol parameters determine the number of rows and number of columns, respectively. When only one of them is specified, the other is automatically determined based on the length of the data vector:
matrix(1:6, nrow = 2)
## [,1] [,2] [,3]
## [1,] 1 3 5
## [2,] 2 4 6
matrix(1:6, ncol = 3)
## [,1] [,2] [,3]
## [1,] 1 3 5
## [2,] 2 4 6matrix(1:6, nrow = 3)
## [,1] [,2]
## [1,] 1 4
## [2,] 2 5
## [3,] 3 6
matrix(1:6, ncol = 2)
## [,1] [,2]
## [1,] 1 4
## [2,] 2 5
## [3,] 3 6Create a
matrixwith 3 rows and 4 columns which contains the numbers 12-1 in decreasing order.
What do you think will happen when we try to create a matrix with less or more
datavalues than matrix sizenrow*ncol? Run the following expressions to find out.
Create a \(3\times3\) matrix where all values are \(1/9\).
5.1.3 matrix properties
5.1.3.1 Dimensions
The length function returns the number of values in a matrix:
The nrow and ncol functions return the number of rows and columns in a matrix, respectively:
The dim function gives both dimensions of the matrix as a vector of length 2, i.e., number of rows and columns, respectively:
For example, the built-in matrix named volcano contains elevation data. Let’s check its length and dimensions:
5.1.3.2 Row and column names
The rownames and colnames functions return the matrix row and column names, respectively. Unlike data.frame row and column names, which are mandatory (Section 4.1.4.2), matrix row and column names are optional. The matrix row and column names can be initialized or modified by assignment to these properties:
5.1.4 matrix conversions
5.1.4.1 matrix → vector
The as.vector function converts a matrix to a vector:
Note that the matrix values are always arranged by column in the resulting vector!
5.1.4.2 matrix → data.frame
The as.data.frame function converts a matrix to a data.frame:
Note that row and column names are automatically generated as part of the conversion, since they are mandatory in a data.frame (Section 5.1.3.2).
5.1.5 Transposing a matrix
The t function transposes a matrix. In other words, the matrix rows and columns are “switched”—rows become columns and columns become rows:
What will be the result of
t(t(x))?
5.1.6 Image with contours
Using the image and contour functions we can graphically display matrix values. The color scale can be set with col and the x/y aspect ratio can be set with asp. Also, add=TRUE is used so that the contour is added on top of the existing plot, rather than initiated in a new plot. The resulting image is shown in Figure 5.1:
image(volcano, col = terrain.colors(30), asp = ncol(volcano) / nrow(volcano))
contour(volcano, add = TRUE)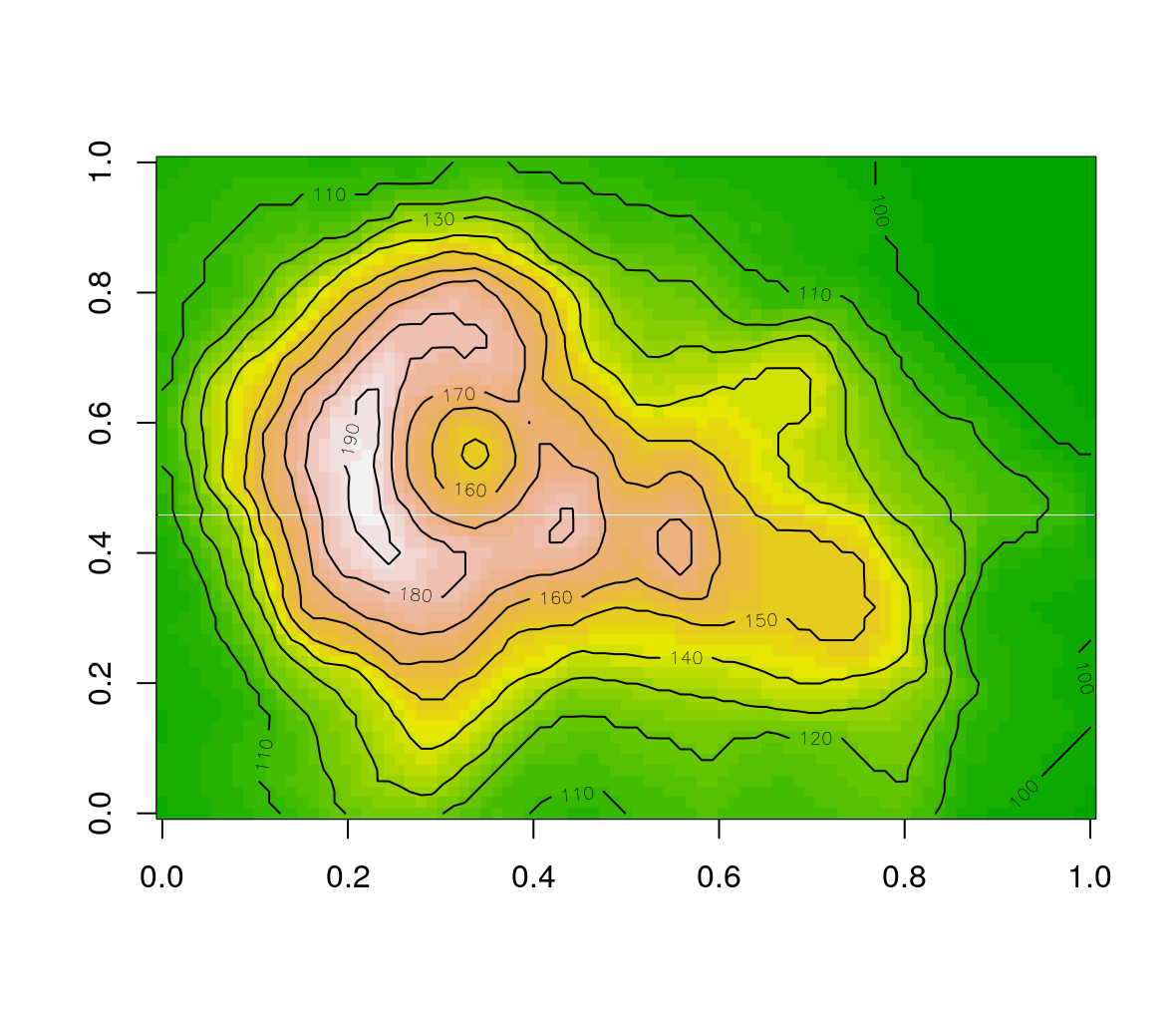
Figure 5.1: Volcano image with contours
5.1.7 Matrix subsetting
5.1.7.1 Individual rows and columns
Similarly to what we learned about data.frame, matrix indices are two-dimensional. The first value refers to rows and the second value refers to columns. For example:
The following examples subset the volcano matrix:
Does the
volcanomatrix contain anyNAvalues? How can we make sure?
Complete rows or columns can be accessed by leaving a blank space instead of the row or column index. By default, a subset that comes from a single row or a single column is simplified to a vector:
To “suppress” the simplification of individual rows/columns to a vector, we can use the drop=FALSE argument (Section 4.1.5.3):
When referring to an elevation matrix, such as volcano, any given row or column subset is actually an elevation profile. For example, the following expressions extract two elevation profiles from volcano:
Figure 5.2 graphically displays those profiles:
plot(r30, type = "o", col = "blue", ylim = range(c(r30, r70)), ylab = "Elevation (m)")
lines(r70, type = "o", col = "red")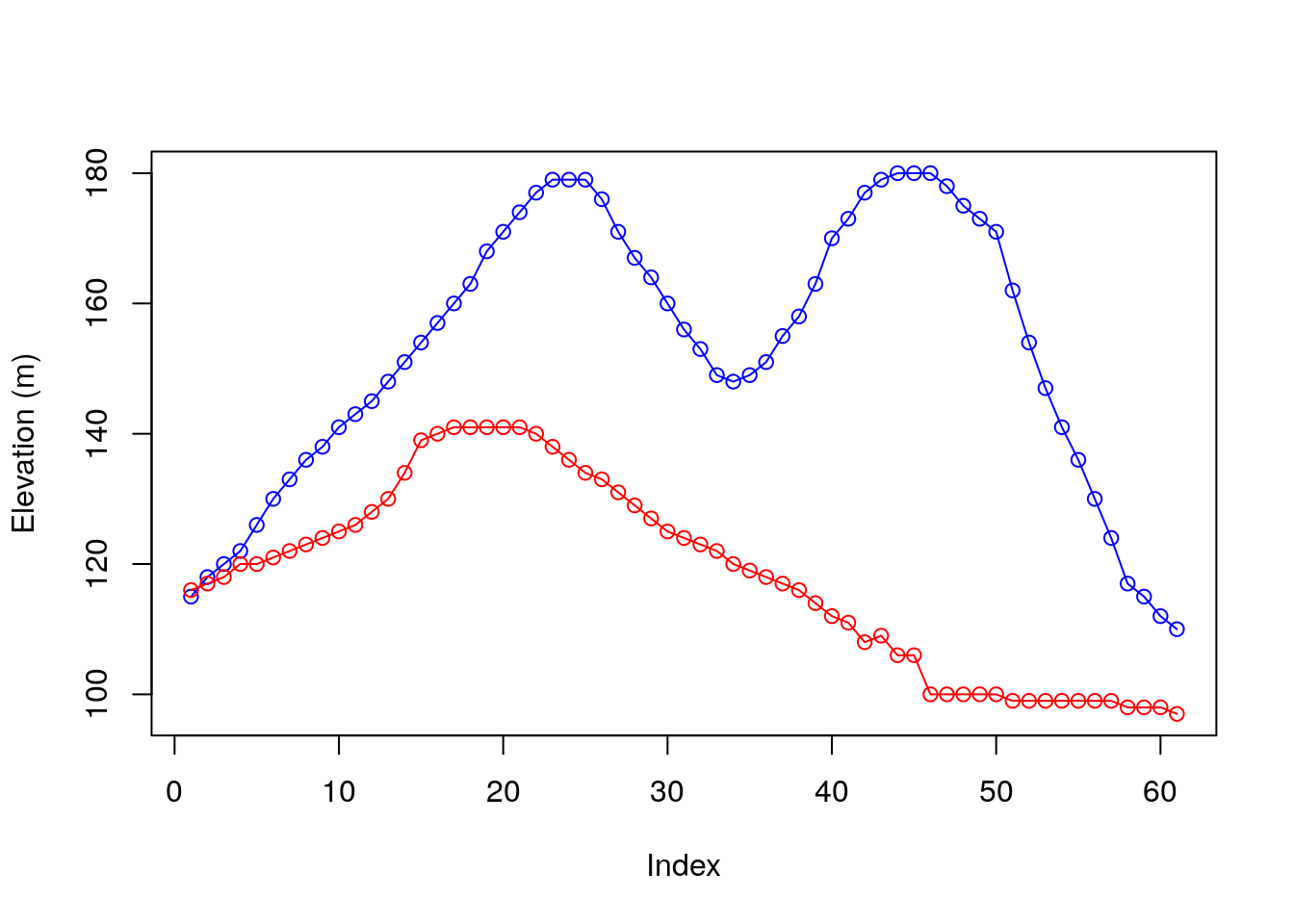
Figure 5.2: Rows 30 (blue) and 70 (red) from the volcano matrix
Figure 5.3 shows the location of the two profiles in a 3D image of volcano.
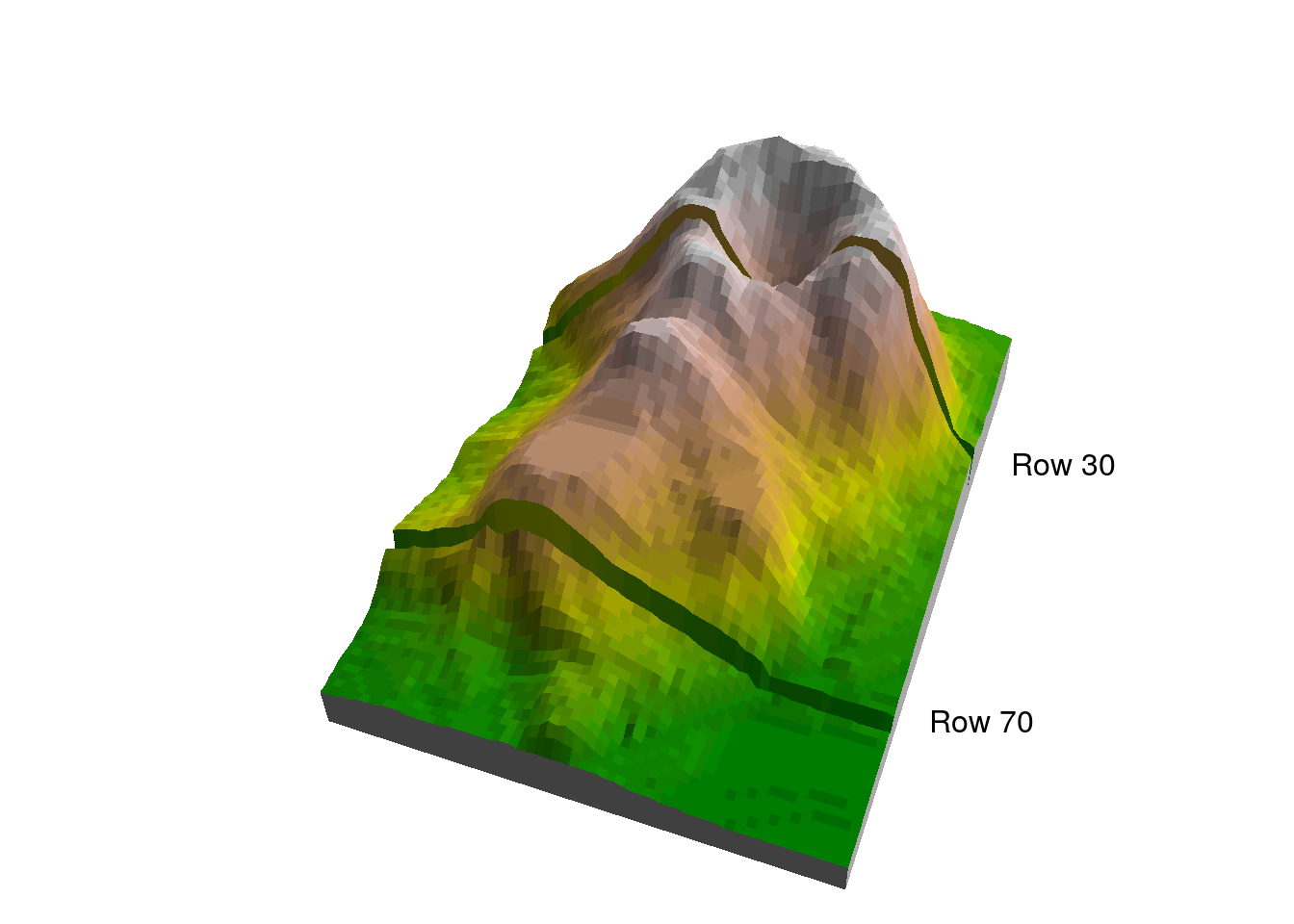
Figure 5.3: Rows 30 and 70 in the volcano matrix
5.1.7.2 Subsets with >1 row or column
We can also use vectors of length >1 when subsetting:
We can assign new values to subsets of a matrix:
5.1.8 Summarizing rows and columns
How can we calculate the row or column means of a matrix? One way is to use a for loop (Section 4.2.3), as follows:
The resulting vector of row means can be visualized as follows (Figure 5.4):
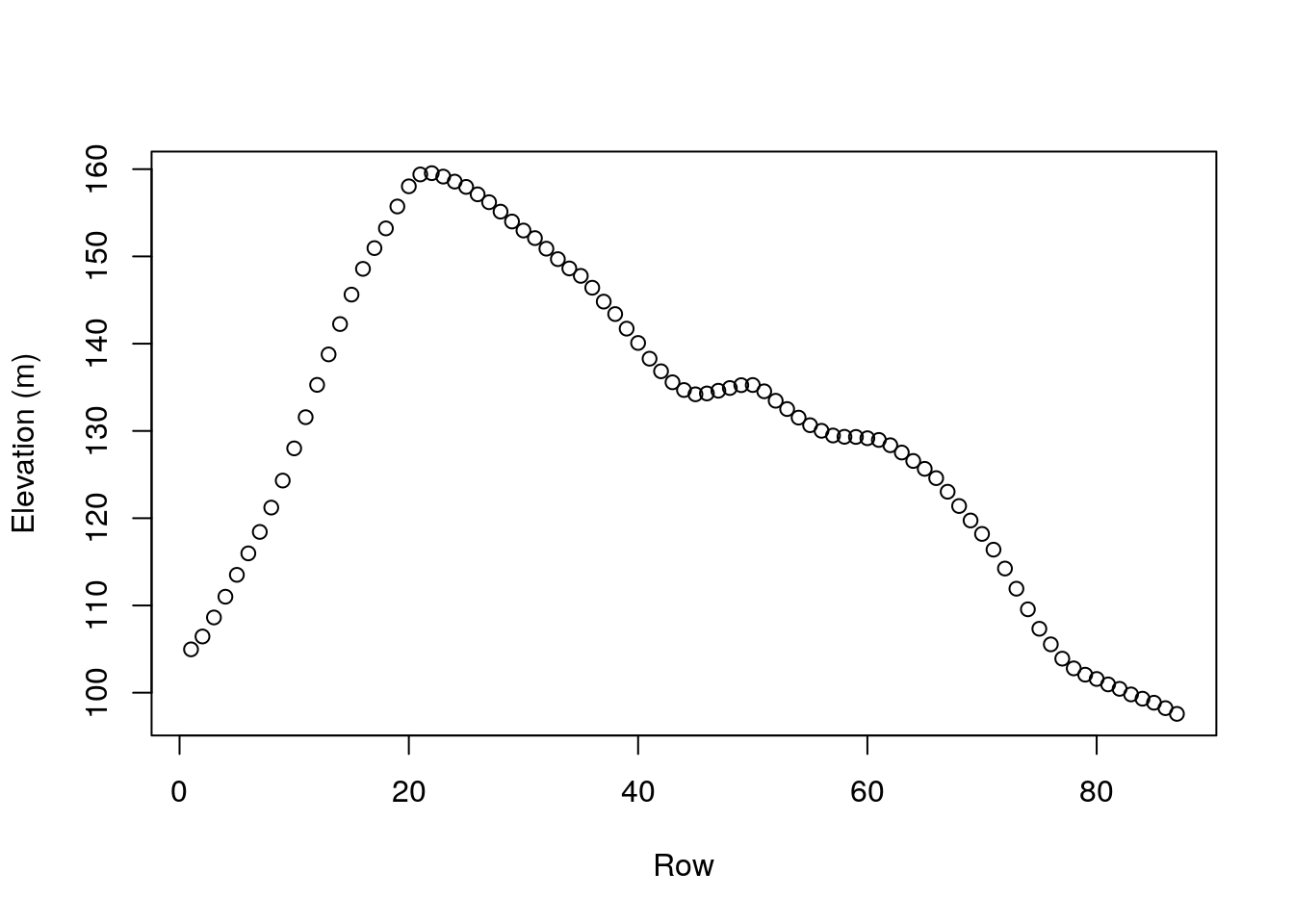
Figure 5.4: Row means of volcano
What changes do we need to make in the code to calculate column means?
We can use the apply function (Section 4.5) to do the same, using much shorter code:
For the special case of mean there are further shortcuts, named rowMeans and colMeans17:
Note: in both cases we can use na.rm to determine whether NA values are included in the calculation (default is FALSE).
How can we check whether the above two expressions give exactly the same result?
5.2 Arrays
5.2.1 Creating an array
An array is a data structure that contains values of the same type and can have any number of dimensions. We may therefore consider a vector (1 dimension) and a matrix (2 dimensions) as special cases of an array.
An array can be created with the array function, specifying the values and the required dimensions. For example, the following expression creates an array with the values 1:24 and three dimensions—2 rows, 3 columns and 4 “layers” (Figure 5.5):
y = array(data = 1:24, dim = c(2, 3, 4))
y
## , , 1
##
## [,1] [,2] [,3]
## [1,] 1 3 5
## [2,] 2 4 6
##
## , , 2
##
## [,1] [,2] [,3]
## [1,] 7 9 11
## [2,] 8 10 12
##
## , , 3
##
## [,1] [,2] [,3]
## [1,] 13 15 17
## [2,] 14 16 18
##
## , , 4
##
## [,1] [,2] [,3]
## [1,] 19 21 23
## [2,] 20 22 24
Figure 5.5: An array with 2 rows, 3 columns and 4 “layers”
5.2.2 Array subsetting
Subsetting an array works similarly to matrix subsetting (Section 5.1.7), only that we can have any number of indices—corresponding to the number of array dimensions—rather than two. Accordingly, when subsetting a three-dimensional array we need to provide three indices. For example, here is how we can extract a particular row, column or “layer” from a three-dimensional array (Figure 5.6):
Figure 5.6: array subsetting: selecting one row, column or “layer”
We can also subset two or three dimensions at a time, to get an individual row/column, row/layer, column/layer or row/column/layer combination (Figure 5.7):
Figure 5.7: array subsetting
5.2.3 Using apply with arrays
When using apply on a 3-dimensional array, we can apply a function:
- On one of the dimensions
- On a combinations of any two dimensions
Here are the four most useful dimension combinations with respect to arrays representing spatial data (Section 5.3):
5.2.4 Basic data structures in R
So far we met four out of five of the basic data structures in R (Table 5.1), so it is time for a short summary. We can classify the basic data structures in R based on:
- Number of dimensions—One-dimensional, two-dimensional or n-demensional18
- Homogeneity—homogeneous (values of the same type) or heterogeneous (values of different types)
| Number of dimensions | Homogeneous | Heterogeneous |
|---|---|---|
| one-dimensional | vector | list |
| two-dimensional | matrix |
data.frame |
| n-dimensional | array |
Most of the data structures in R are combinations of the basic five ones.
5.3 Rasters
5.3.1 What is a raster?
A raster (Figure 5.8) is basically a matrix or an array, representing a rectangular area on the surface of the earth. To associate the matrix or the array with the particular area it represents, the raster has some additional spatial properties, on top of the non-spatial properties that any ordinary matrix or array has:
- Non-spatial properties
- Values
- Dimensions (rows, columns, layers)
- Spatial properties
- Extent
- Coordinate Reference System (CRS)
- (Resolution)
![Raster cells^[http://desktop.arcgis.com/en/arcmap/10.3/manage-data/raster-and-images/what-is-raster-data.htm]](images/lesson_05_raster_esri.png)
Figure 5.8: Raster cells19
Raster extent is the range of x- and y-axis coordinates that the raster occupies (Figure 5.9). The Coordinate Reference System (CRS) is the particular system that “associates” the raster coordinates (which are just pairs of x/y values) to geographic locations. Raster resolution is the size of a raster cell, in the x and y directions. The resolution is listed in parentheses because it can be calculated given the extent and the number of rows and columns. For example, the x-axis resolution is equal to the x-axis range difference (i.e., \(x_{max} - x_{min}\)) divided by the number of columns. In the leftmost panel in Figure 5.9, the the x-axis and y-axis resolutions are both equal to \(\frac{8-0}{12}=\frac{8}{12}\approx 0.67\).
![Rasters with the same extent but four different resolutions^[http://datacarpentry.org/organization-geospatial/01-intro-raster-data/index.html]](images/lesson_05_datacarpentry_raster_resolution.png)
Figure 5.9: Rasters with the same extent but four different resolutions20
5.3.2 Raster file formats
Commonly used raster file formats (Table 5.2) can be divided in two groups. “Simple” raster file formats, such as GeoTIFF, are single-band or multi-band rasters (Figure 5.10) where the extent is geo-referenced, as discussed above (Section 5.3.1). “Complex” raster file formats, such as HDF, contain additional complexity, such as more than three dimensions (Figure 5.11, see Section 5.3.9 below) and/or metadata, such as band names, time stamps, units of measurement, and so on.
| Type | Format | File extension |
|---|---|---|
| “Simple” | GeoTIFF | .tif |
| Erdas Imagine Image | .img |
|
| “Complex” (>3D and/or metadata) | HDF | .hdf |
| NetCDF | .nc |
![Single-band and multi-band raster^[https://datacarpentry.org/organization-geospatial/]](images/lesson_05_raster_single_band_vs_multi_band.png)
Figure 5.10: Single-band and multi-band raster21
![An example of an HDF file structure^[http://matthewrocklin.com/blog/work/2018/02/06/hdf-in-the-cloud]](images/HDF_structure.png)
Figure 5.11: An example of an HDF file structure22
5.3.3 Using R packages
An R package is a collection of code files used to load objects—mostly functions—into memory. All object definitions in R are contained in packages. To use a particular object, we need to:
- Install the package with the
install.packagesfunction (once) - Load the package using the
libraryfunction (in each new R session)
Loading a package with library basically means that all of its code files are executed, loading all objects the package defined into the RAM. However, all function calls we used until now did require installing or loading a package. Why is that? Because several packages are installed along with R and loaded on R start-up. There are several more packages which are installed by default but not loaded on start-up (total of ~30) (Figure 5.12).
Figure 5.12: Packages included with R23
Most of the ~15,000 R packages (as of October 2019) are not installed by default. To use one of these packages we first need to install it on the computer. Installing a package is a one-time operation using the install.packages function. After the package is installed, each time we want to use it we need to load it using library.
In the following examples, we are going use a package called stars to work with rasters. The stars package is not installed with R, therefore we need to install it ourselves with:
If the package is already installed, running install.packages overwrites the old installation. This is done intentionally if you you want to install a newer version of the package. Once the package is already installed, we need to use the library function to load it into memory. Note how package name can be passed to library without quotes:
Other than stars, we are going to use the mapview, cubeview, sf, units, gstat and automap packages.
5.3.4 The raster package
Before moving on to stars package, the raster package deserves to be mentioned. The raster package is a powerful and well-established (2010-) package for working with rasters in R (Table 0.1). This package contains several classes and functions for working with rasters in R. The most important three classes from raster are the ones for representing rasters:
RasterLayerfor single-band rastersRasterStackandRasterBrickfor multi-band rasters24
The raster package has several limitations, most notably that it is limited to three dimensions and cannot hold raster metadata, such as layer names and measurement units. Moreover, it is not well-integrated with the sf package for vector layers. For those reasons, in this book we will be working with the newer stars package for rasters (Section 5.3.5).
5.3.5 The stars package
The stars package is a newer (2018-) R package for working with rasters (Table 0.1). The stars package is more general and more tighly integrated with vector layer analysis in the sf package, compared to raster.
The stars package contains the stars class for representing all types of rasters, and numerous functions for working with rasters in R. A stars object is basically a list of matrices or arrays, along with metadata describing their dimensions. Don’t worry if this is not clear; we will elaborate on this later on (Section 6.3).
Compared to the three raster classes (Section 5.3.4), the stars class is more flexible and can represent more complex types of rasters:
- Rasters with more than three dimensions
- Metadata (band names, units of measurement, etc.)
- Not-standard grids (rotated, etc.)
5.3.6 Reading raster from file
The most common and most useful method for creating a raster object in R (and elsewhere) is reading from a file, such as a GeoTIFF file. We can use the read_stars function to read a GeoTIFF file and create a stars object in R. The first parameter (.x) is the file path to the file we want to read, or just the file name—when the file is located in the working directory.
As an example, let’s read the dem.tif raster file, which contains a coarse Digital Elevation Model (DEM) of the area around Haifa. First, we have to load the stars package:
Then, we can use the read_stars function to read the file25:
The file MOD13A3_2000_2019.tif is a multi-band raster with monthly Normalized Difference Vegetation Index (NDVI) values in Israel, for the period between February 2000 and June 2019 from the MODIS instrument on the Terra satellite. This is a multi-band raster with 233 bands, where each band corresponds to an average monthly NDVI image (233 months total). Uncertain values were replaced with NA. Let’s try reading this file as well, to create another stars object in the R environment:
Two stars objects, named r and s, are now available in our R session.
5.3.7 Visualization with plot, mapview and cubeview
5.3.7.1 Raster images with plot
The simplest way to visualize a stars object is to use the plot function. This produces a static image of the raster, such as the ones shown in Figures 5.13 and 5.14:
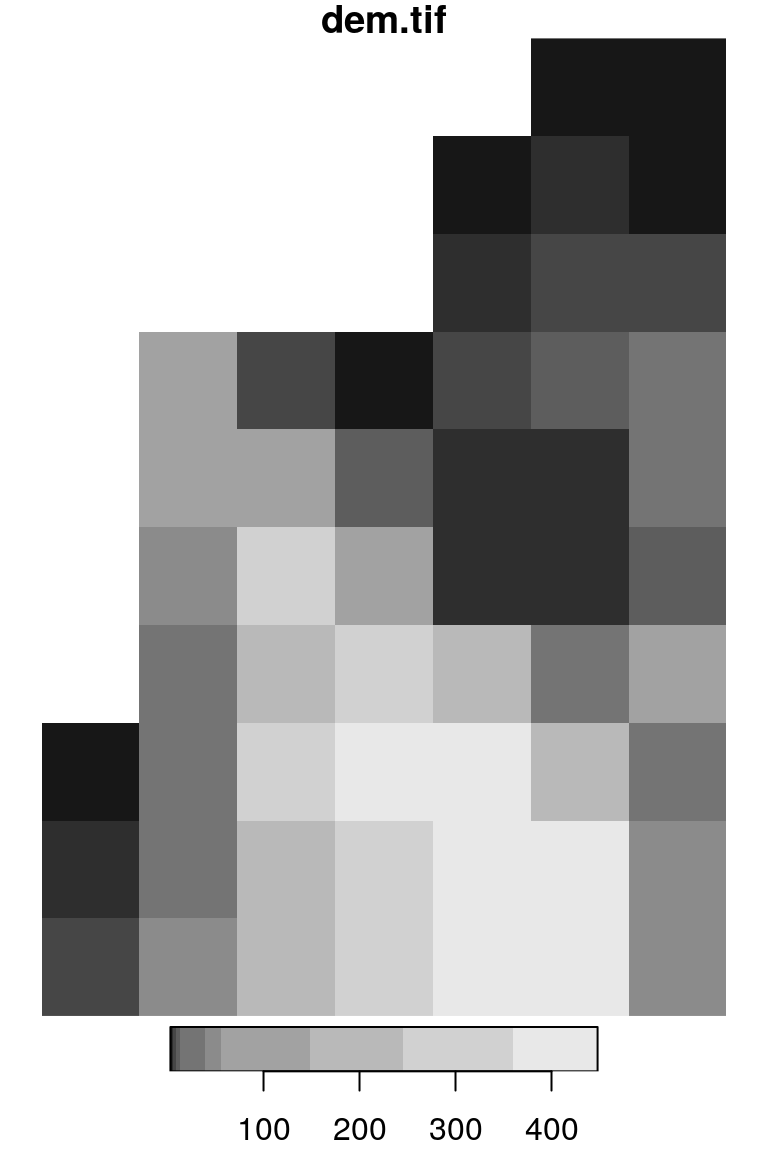
Figure 5.13: A default raster plot output: single-band raster
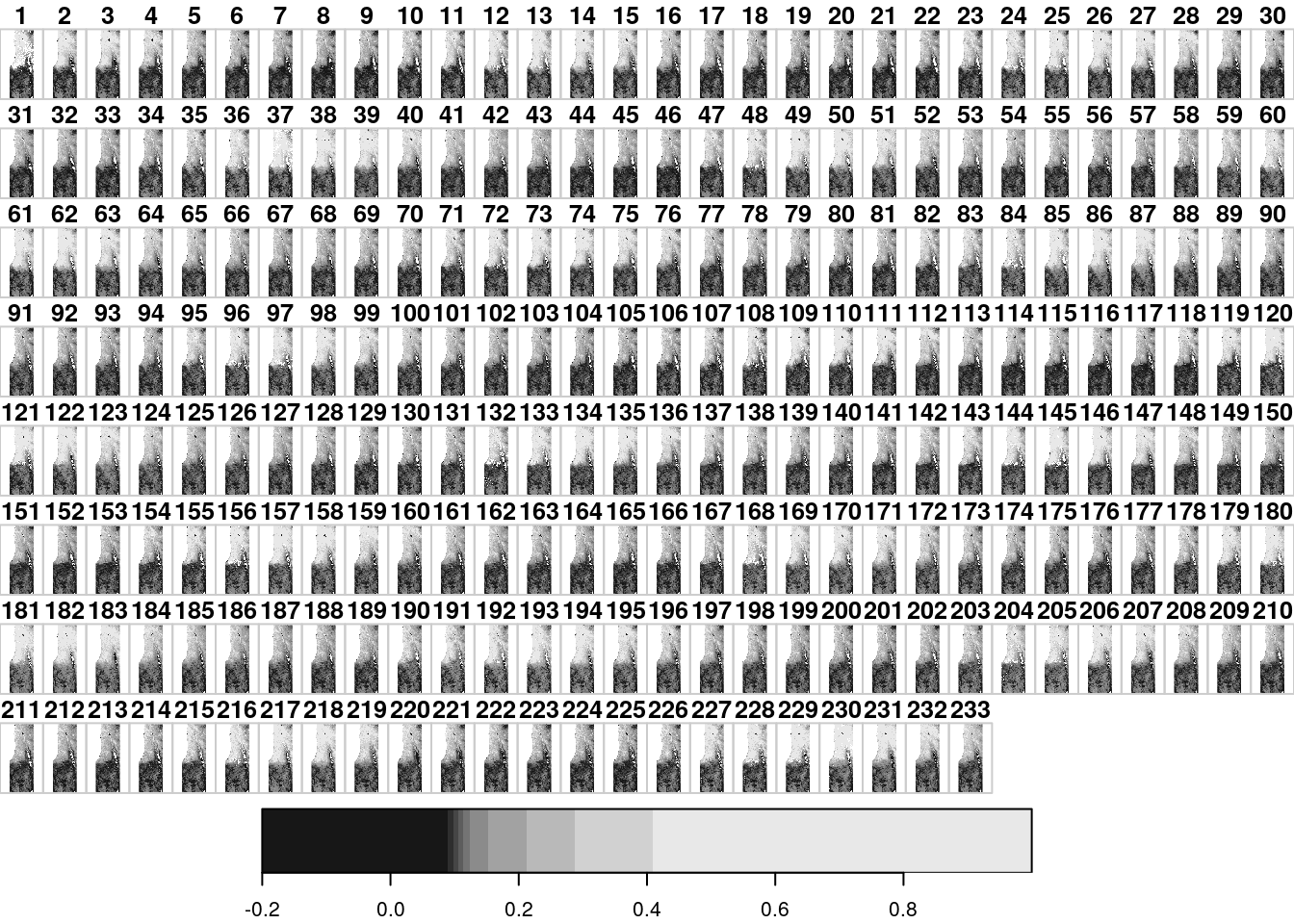
Figure 5.14: A default raster plot output: multi-band raster
Useful additional parameters when running plot on stars objects include:
text_values—Logical, whether to display text labelsaxes—Logical, whether to display axescol—A vector of color codes or names
For example (Figure 5.15):
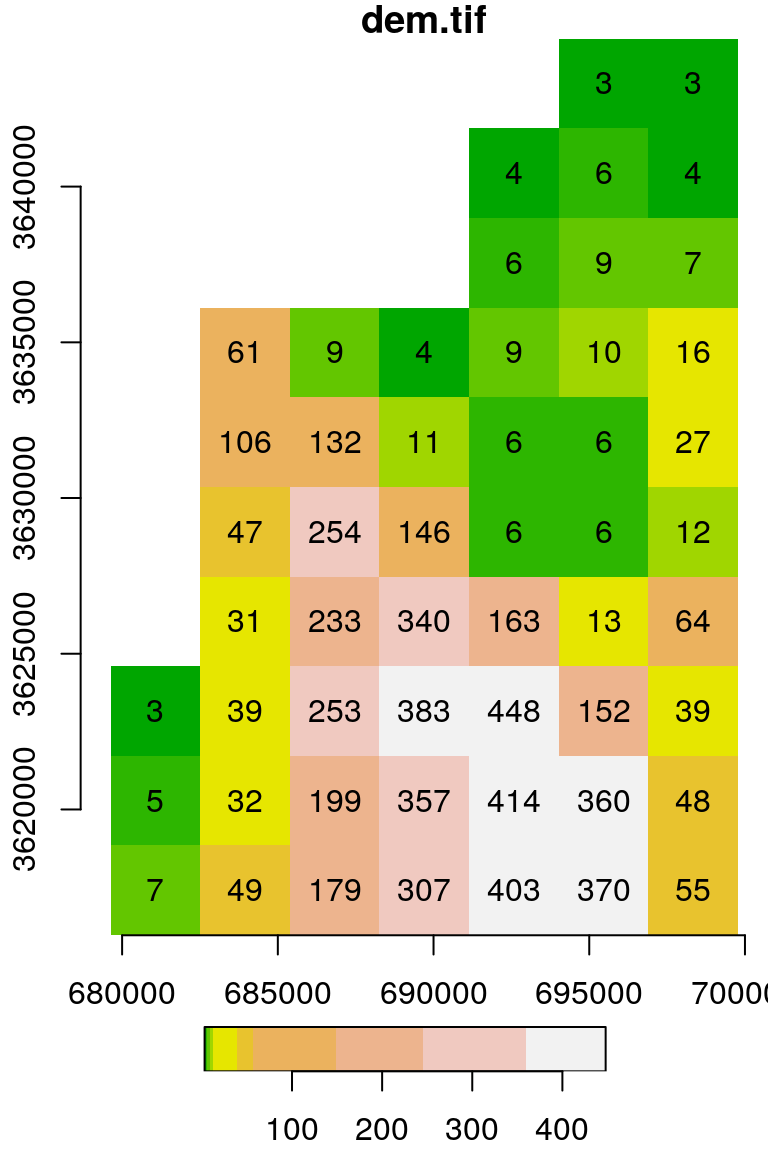
Figure 5.15: Raster plot with additional text_values, axes and col settings
The expression terrain.colors(10) uses one of the built-in color palette functions in R to generate a vector of length 10 with terrain color codes:
terrain.colors(10)
## [1] "#00A600" "#2DB600" "#63C600" "#A0D600" "#E6E600" "#E8C32E" "#EBB25E"
## [8] "#EDB48E" "#F0C9C0" "#F2F2F2"The default color breaks are calculated using quantiles (breaks="quantile"). We can use other break types (such as breaks="equal") or pass our own vector of custom breaks (Figure 5.16):
plot(s, text_values = TRUE, col = terrain.colors(10), breaks = "equal")
plot(s, text_values = TRUE, col = terrain.colors(3), breaks = c(0, 100, 300, 500))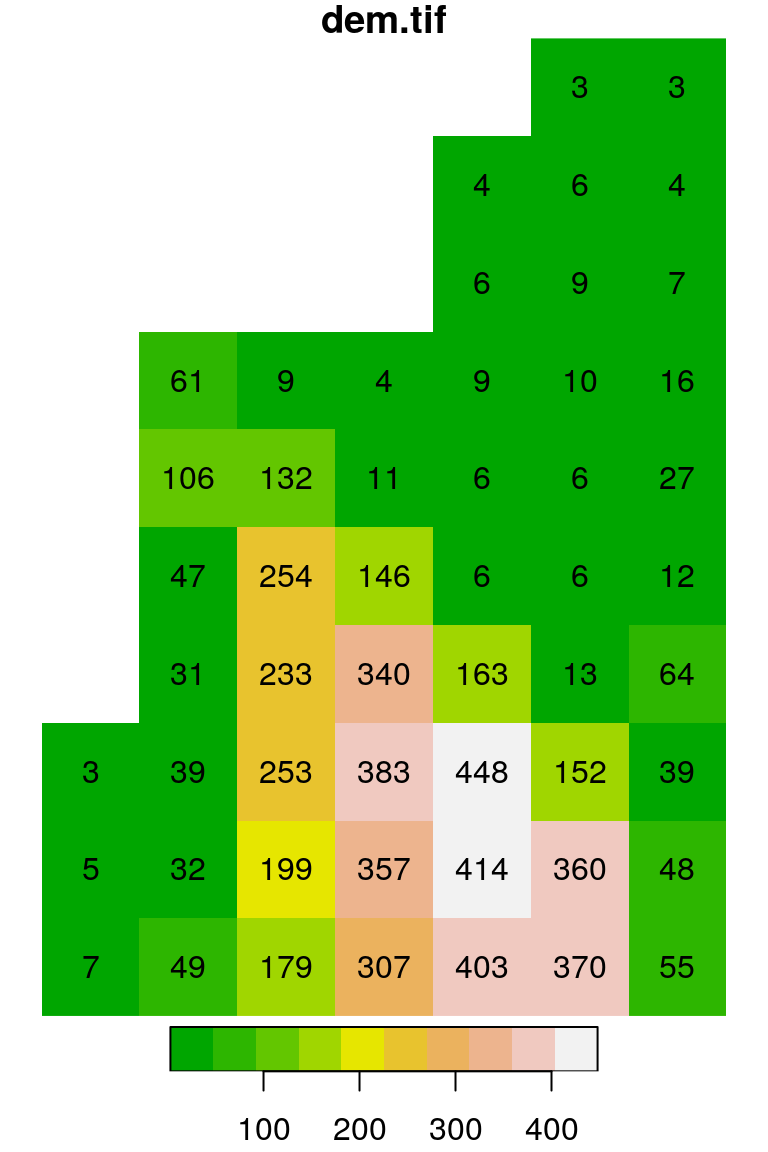
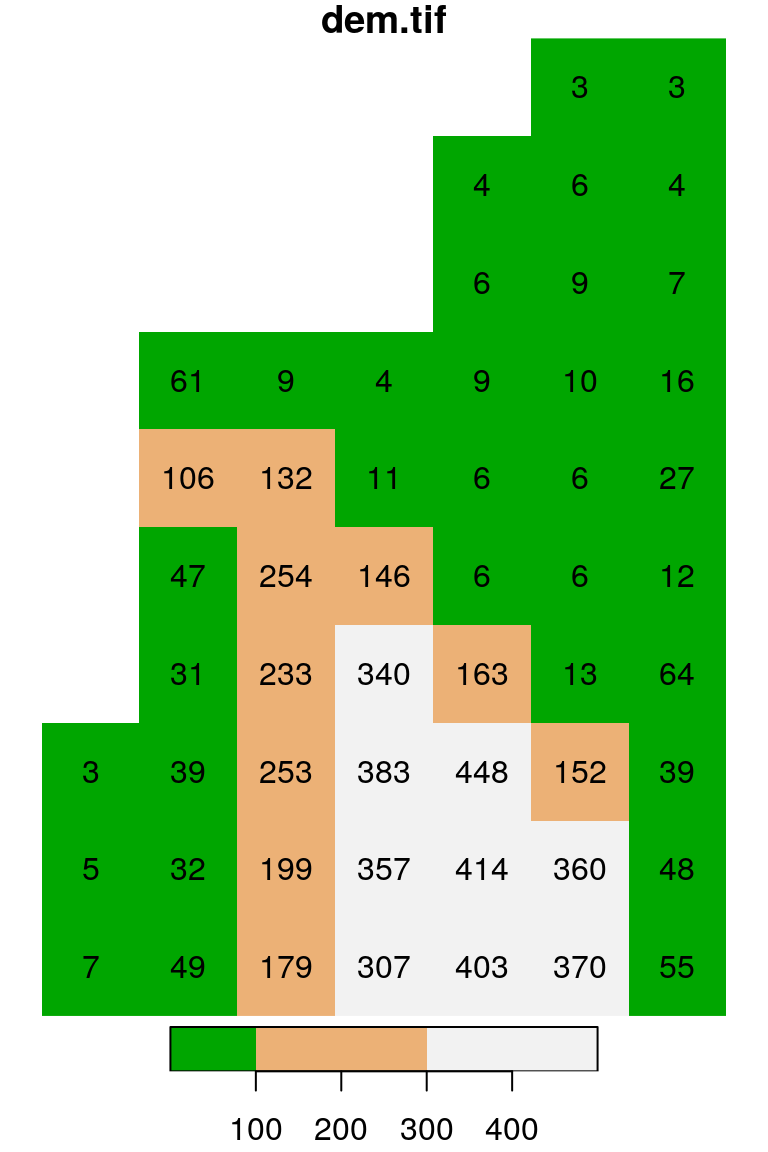
Figure 5.16: Evenly spaced color breaks using breaks = "equal" (left) and manually defined breaks using breaks = c(0, 100, 300, 500) (right)
Note that the number of colors in the second expression (3) matches the number of breaks minus 1 (why?).
5.3.7.2 Interactive maps with mapview
The mapview function from package mapview lets us visually examine spatial objects—vector layers or rasters—in an interactive map on top of various background layers, such as OpenStreetMap, satellite images, etc. We can use the mapview function, after loading the mapview package, as follows:
5.3.7.3 Interactive cubes with cubeview
Another useful package for examining rasters, specifically multi-band ones, is cubeview (Figure 5.17):
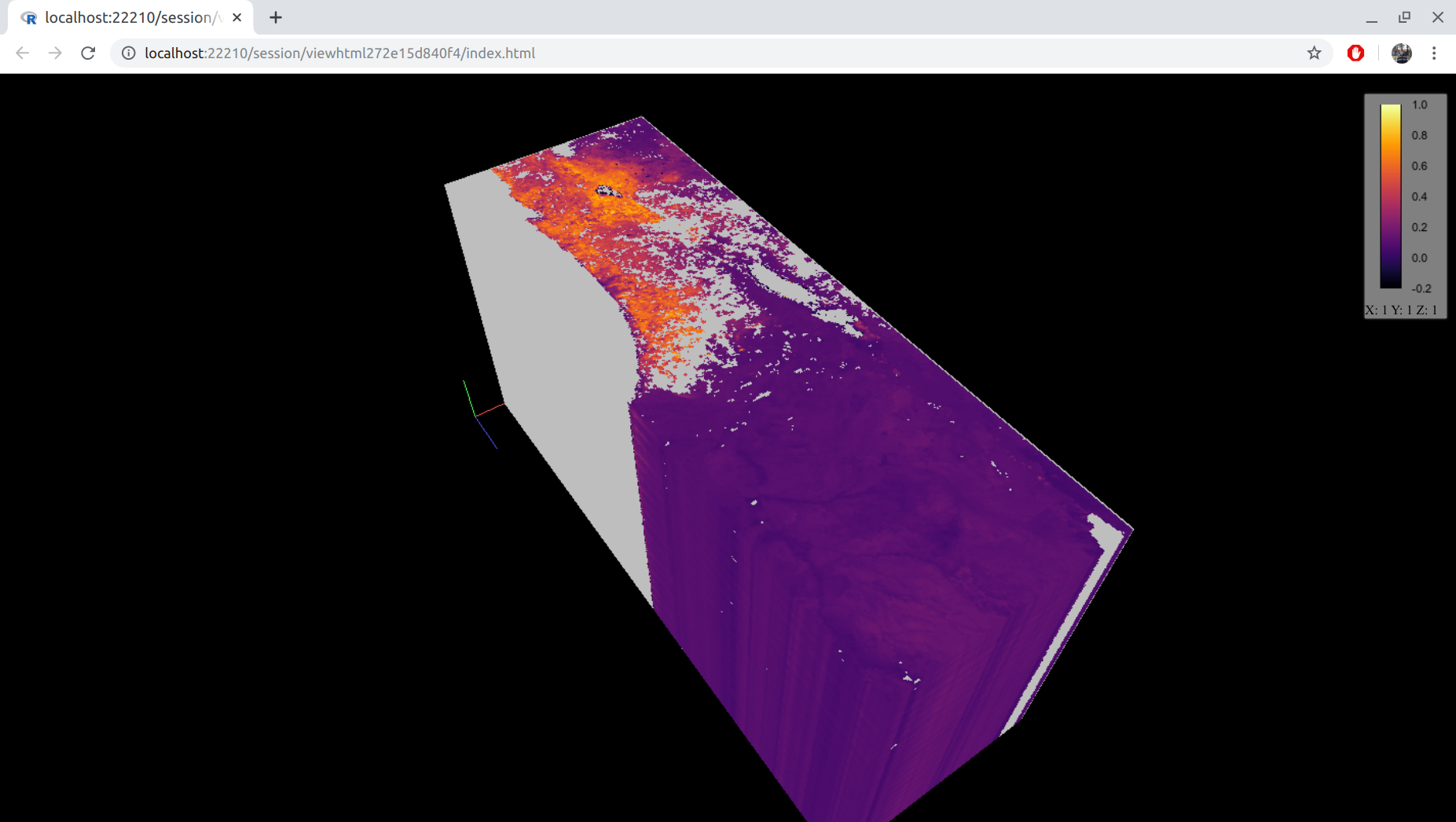
Figure 5.17: A three-dimensional raster visualized with cubeview
5.3.8 Raster values and properties
5.3.8.1 Class and structure
The print method for raster objects gives a summary of their properties:
s
## stars object with 2 dimensions and 1 attribute
## attribute(s):
## dem.tif
## Min. : 3.0
## 1st Qu.: 7.0
## Median : 39.0
## Mean :110.5
## 3rd Qu.:179.0
## Max. :448.0
## NA's :17
## dimension(s):
## from to offset delta refsys point values
## x 1 7 679624 2880 UTM Zone 36, Northern Hem... FALSE NULL [x]
## y 1 10 3644759 -2880 UTM Zone 36, Northern Hem... FALSE NULL [y]The class function returns the class name, which is stars in this case:
As discussed in Section 1.1.5, a class is a “template” with pre-defined properties that each object of that class has. For example, a stars object is actially a collection (namely, a list) of matrix or array objects, along with additional properties of the dimensions, such as dimension names, Coordinate Reference Systems (CRS), etc. When we read the dem.tif file with read_stars, the information regarding all of the properties was transferred from the file and into the stars “template”. Now, the stars object named s is in the RAM, filled with the specific values from dem.tif.
We can display the structure of the stars object with the specific values with str:
str(s)
## List of 1
## $ dem.tif: num [1:7, 1:10] NA NA NA NA NA 3 3 NA NA NA ...
## - attr(*, "dimensions")=List of 2
## ..$ x:List of 7
## .. ..$ from : num 1
## .. ..$ to : num 7
## .. ..$ offset: num 679624
## .. ..$ delta : num 2880
## .. ..$ refsys:List of 2
## .. .. ..$ input: chr "UTM Zone 36, Northern Hemisphere"
## .. .. ..$ wkt : chr "PROJCRS[\"UTM Zone 36, Northern Hemisphere\",\n BASEGEOGCRS[\"WGS 84\",\n DATUM[\"unknown\",\n "| __truncated__
## .. .. ..- attr(*, "class")= chr "crs"
## .. ..$ point : logi FALSE
## .. ..$ values: NULL
## .. ..- attr(*, "class")= chr "dimension"
## ..$ y:List of 7
## .. ..$ from : num 1
## .. ..$ to : num 10
## .. ..$ offset: num 3644759
## .. ..$ delta : num -2880
## .. ..$ refsys:List of 2
## .. .. ..$ input: chr "UTM Zone 36, Northern Hemisphere"
## .. .. ..$ wkt : chr "PROJCRS[\"UTM Zone 36, Northern Hemisphere\",\n BASEGEOGCRS[\"WGS 84\",\n DATUM[\"unknown\",\n "| __truncated__
## .. .. ..- attr(*, "class")= chr "crs"
## .. ..$ point : logi FALSE
## .. ..$ values: NULL
## .. ..- attr(*, "class")= chr "dimension"
## ..- attr(*, "raster")=List of 3
## .. ..$ affine : num [1:2] 0 0
## .. ..$ dimensions : chr [1:2] "x" "y"
## .. ..$ curvilinear: logi FALSE
## .. ..- attr(*, "class")= chr "stars_raster"
## ..- attr(*, "class")= chr "dimensions"
## - attr(*, "class")= chr "stars"The unclass function removes the class definition from an R object. This is another convenient way to demonstrate the fact that a stars object s is composed of a numeric matrix along with the spatial properties of the x and y dimensions:
unclass(s)
## $dem.tif
## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## [1,] NA NA NA NA NA NA NA 3 5 7
## [2,] NA NA NA 61 106 47 31 39 32 49
## [3,] NA NA NA 9 132 254 233 253 199 179
## [4,] NA NA NA 4 11 146 340 383 357 307
## [5,] NA 4 6 9 6 6 163 448 414 403
## [6,] 3 6 9 10 6 6 13 152 360 370
## [7,] 3 4 7 16 27 12 64 39 48 55
##
## attr(,"dimensions")
## from to offset delta refsys point values
## x 1 7 679624 2880 UTM Zone 36, Northern Hem... FALSE NULL [x]
## y 1 10 3644759 -2880 UTM Zone 36, Northern Hem... FALSE NULL [y]5.3.8.2 Raster attributes and values
stars objects are collections of matrices or arrays. Each matrix or array is known as an attribute and is associated with a name. A GeoTIFF file, being a simple raster format (Section 5.3.2), can contain just one attribute. Attribute names are not specified as part of a GeoTIFF file, and therefore automatically given default values based on the file name. We can get the attribute name(s) using the names function:
We can change the attribute names through assignment. For example, it makes sense to name the attribute after the physical property or measurement it represents:
Accessing an attrubute, by name or by numeric index, returns the matrix (single-band raster) or array (multi-band raster) object with the values of that attribute. For example, in the following expressions we access the (only) attribute of the s and r rasters by name:
s$elevation
## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## [1,] NA NA NA NA NA NA NA 3 5 7
## [2,] NA NA NA 61 106 47 31 39 32 49
## [3,] NA NA NA 9 132 254 233 253 199 179
## [4,] NA NA NA 4 11 146 340 383 357 307
## [5,] NA 4 6 9 6 6 163 448 414 403
## [6,] 3 6 9 10 6 6 13 152 360 370
## [7,] 3 4 7 16 27 12 64 39 48 55
class(s$elevation)
## [1] "matrix" "array"
dim(s$elevation)
## x y
## 7 10and here we do the same using a numeric index and the [[ operator:
The $ and [[ operators actually select individual elements from a list; this works because a stars object is, internally, a list of matrices or arrays (i.e., the “attributes”). We will elaborate on lists and the [[ operator in Section 11.1.
By now we met all three subset operators which we are going to use in this book (Table 5.3).
| Syntax | Objects | Returns |
|---|---|---|
x[i] |
vector, table, matrix, array, list | Subset i |
x[[i]] |
vectors, lists | Single element i |
x$i |
tables, lists | Single element i |
x@n |
S4 objects | Slot n |
5.3.8.3 Dimensions and spatial properties
The nrow, ncol and dim functions return the number or rows, column or all available dimensions of a stars object, respectively. These functions return a named numeric vector, where the names correspond to dimension names (Section 6.3.2). For example:
As mentioned above (Section 5.3.1), the spatial properties, determining raster placement in geographical space, include the extent, the Coordinate Reference System (CRS) and the resolution.
For example, the CRS definition of a stars object can be accessed with the st_crs function. Here are the CRS definitions of rasters s and r:
st_crs(s)
## Coordinate Reference System:
## User input: UTM Zone 36, Northern Hemisphere
## wkt:
## PROJCRS["UTM Zone 36, Northern Hemisphere",
## BASEGEOGCRS["WGS 84",
## DATUM["unknown",
## ELLIPSOID["WGS84",6378137,298.257223563,
## LENGTHUNIT["metre",1,
## ID["EPSG",9001]]]],
## PRIMEM["Greenwich",0,
## ANGLEUNIT["degree",0.0174532925199433,
## ID["EPSG",9122]]]],
## CONVERSION["Transverse Mercator",
## METHOD["Transverse Mercator",
## ID["EPSG",9807]],
## PARAMETER["Latitude of natural origin",0,
## ANGLEUNIT["degree",0.0174532925199433],
## ID["EPSG",8801]],
## PARAMETER["Longitude of natural origin",33,
## ANGLEUNIT["degree",0.0174532925199433],
## ID["EPSG",8802]],
## PARAMETER["Scale factor at natural origin",0.9996,
## SCALEUNIT["unity",1],
## ID["EPSG",8805]],
## PARAMETER["False easting",500000,
## LENGTHUNIT["metre",1],
## ID["EPSG",8806]],
## PARAMETER["False northing",0,
## LENGTHUNIT["metre",1],
## ID["EPSG",8807]]],
## CS[Cartesian,2],
## AXIS["easting",east,
## ORDER[1],
## LENGTHUNIT["metre",1,
## ID["EPSG",9001]]],
## AXIS["northing",north,
## ORDER[2],
## LENGTHUNIT["metre",1,
## ID["EPSG",9001]]]]st_crs(r)
## Coordinate Reference System:
## User input: unnamed
## wkt:
## PROJCRS["unnamed",
## BASEGEOGCRS["unnamed ellipse",
## DATUM["unknown",
## ELLIPSOID["unnamed",6371007.181,0,
## LENGTHUNIT["metre",1,
## ID["EPSG",9001]]]],
## PRIMEM["Greenwich",0,
## ANGLEUNIT["degree",0.0174532925199433,
## ID["EPSG",9122]]]],
## CONVERSION["Sinusoidal",
## METHOD["Sinusoidal"],
## PARAMETER["Longitude of natural origin",0,
## ANGLEUNIT["degree",0.0174532925199433],
## ID["EPSG",8802]],
## PARAMETER["False easting",0,
## LENGTHUNIT["metre",1],
## ID["EPSG",8806]],
## PARAMETER["False northing",0,
## LENGTHUNIT["metre",1],
## ID["EPSG",8807]]],
## CS[Cartesian,2],
## AXIS["easting",east,
## ORDER[1],
## LENGTHUNIT["metre",1,
## ID["EPSG",9001]]],
## AXIS["northing",north,
## ORDER[2],
## LENGTHUNIT["metre",1,
## ID["EPSG",9001]]]]The CRS definition is an object of class crs, which contains the textual definition of the CRS, in the “proj-strings” format and, when available, also the EPSG code of the CRS.
The extent (“bounding boxes”) can be accessed with st_bbox. Here are the extents of s and r:
The extent is returned as an object of class bbox. The object is basically a numeric vector of length 4, including the xmin, ymin, xmax and ymax values.
The resolution, as well as other properties of a stars dimensions, can be accessed using the st_dimensions function. We will elaborate on this in Section 6.3.1. In the meanwhile, for completeness, here are the resolutions ("delta") of the x- and y-axes of s and r:
Note that the resolution is separate for the "x" and "y" dimensions. The (absolute) resolution is usually equal for both, in which case raster pixels are square. However, the "x" and "y" resolutions can also be unequal, in which case raster pixels are non-square rectangles.
5.3.8.4 Accessing raster values
As shown above (Section 5.3.8.2), raster values can be accessed directly, as a matrix or an array, by selecting a raster attribute, either by name or by index. For example:
s[[1]]
## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## [1,] NA NA NA NA NA NA NA 3 5 7
## [2,] NA NA NA 61 106 47 31 39 32 49
## [3,] NA NA NA 9 132 254 233 253 199 179
## [4,] NA NA NA 4 11 146 340 383 357 307
## [5,] NA 4 6 9 6 6 163 448 414 403
## [6,] 3 6 9 10 6 6 13 152 360 370
## [7,] 3 4 7 16 27 12 64 39 48 55A histogram can give a first impression of the raster values distribution (Figure 5.18), using the hist function:
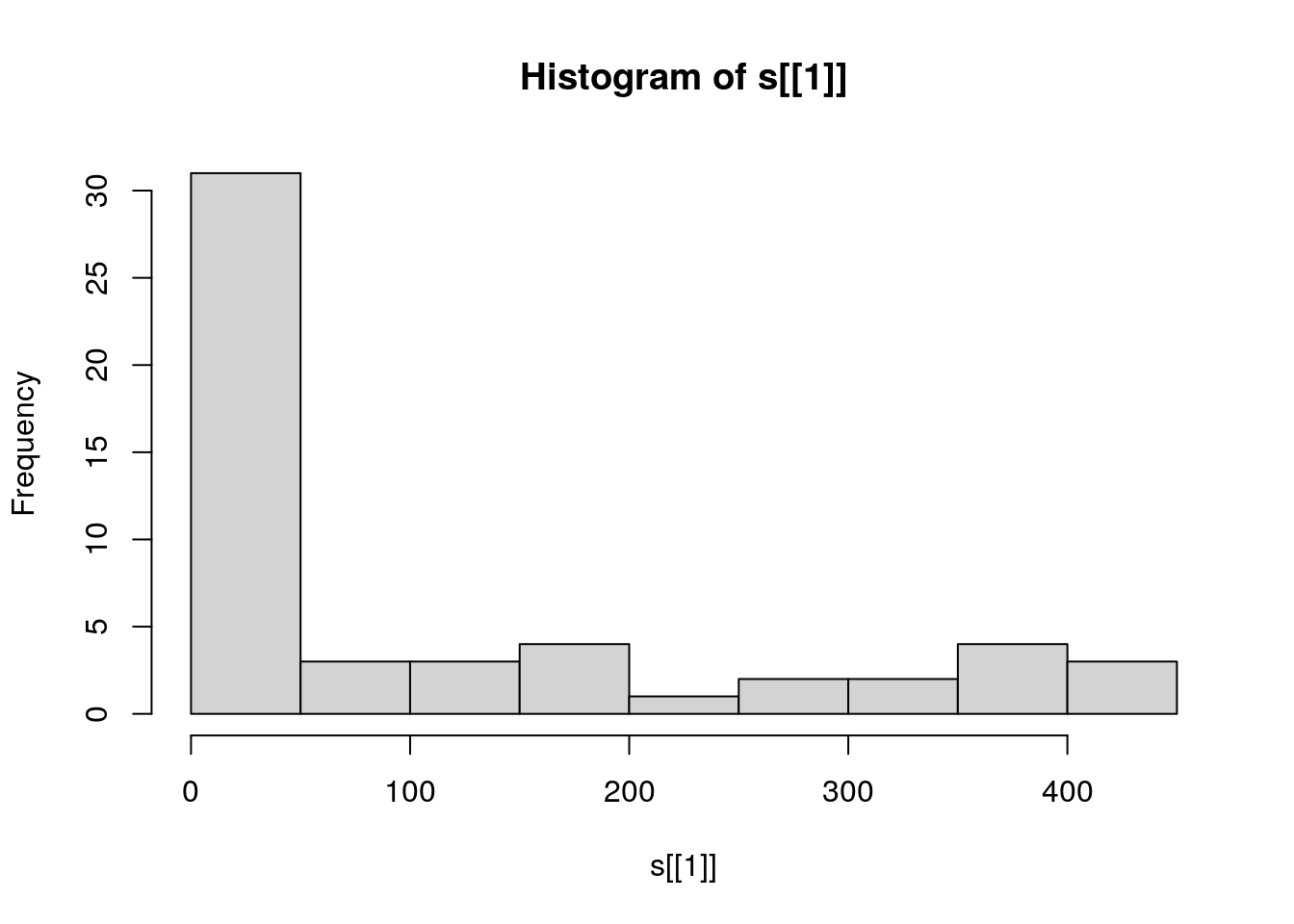
Figure 5.18: Distribution of elevation values
For example, the above histogram tells us that the overall range of elevation values in the raster s is roughly 0-450 (meters), but most pixels are in the 0-50 range.
Note that we can pass the matrix (or array) of raster values to many other functions that accept a numeric vector, such as mean or range.
Calculate the mean, minimum and maximum of the cell values in the raster
s(excludingNA).
The matrix or array rows and columns are reversed compared to the visual arrangement of the raster, because in a stars object the first dimension (matrix rows) refers to x (raster columns) and the second dimension (matrix columns) refers to y (raster rows)! Therefore, for example, the following expression gives the 7th column of the matrix with the raster values, which is actually the 7th row in the raster (Figure 5.15):
We can modify (a subset of) raster values using assignment. For example, the following code creates a copy of the raster s, named u, then replaces the values in the 7th row with the value -1:
The result is shown in Figure 5.19:
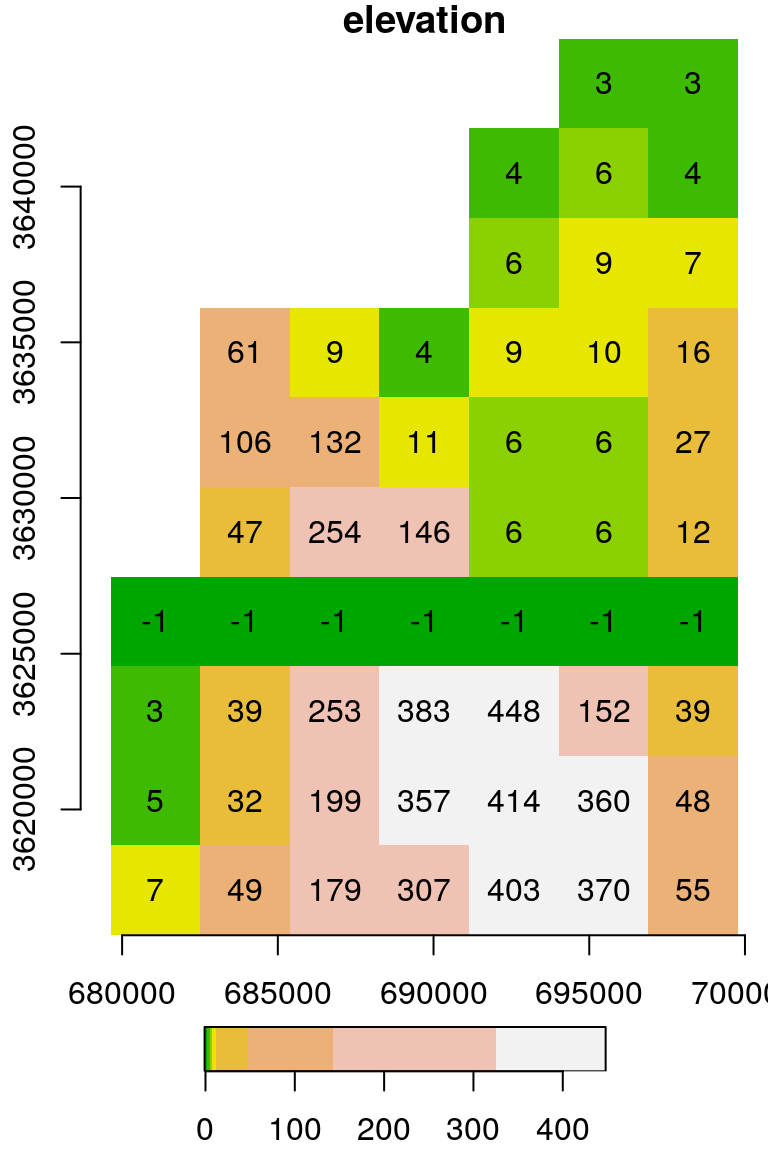
Figure 5.19: Raster with the value -1 assigned into the 7th row
We can even replace the entire matrix or array of values with a custom one. This can be done using assignment to an “empty” subset, which implies selecting all cells, as in r[[1]][]. For example, the following code section creates another copy named u, then replaces all values with a consecutive vector:
u = s
u[[1]][] = 1:length(u[[1]])
u[[1]]
## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## [1,] 1 8 15 22 29 36 43 50 57 64
## [2,] 2 9 16 23 30 37 44 51 58 65
## [3,] 3 10 17 24 31 38 45 52 59 66
## [4,] 4 11 18 25 32 39 46 53 60 67
## [5,] 5 12 19 26 33 40 47 54 61 68
## [6,] 6 13 20 27 34 41 48 55 62 69
## [7,] 7 14 21 28 35 42 49 56 63 70The result is shown in Figure 5.20:
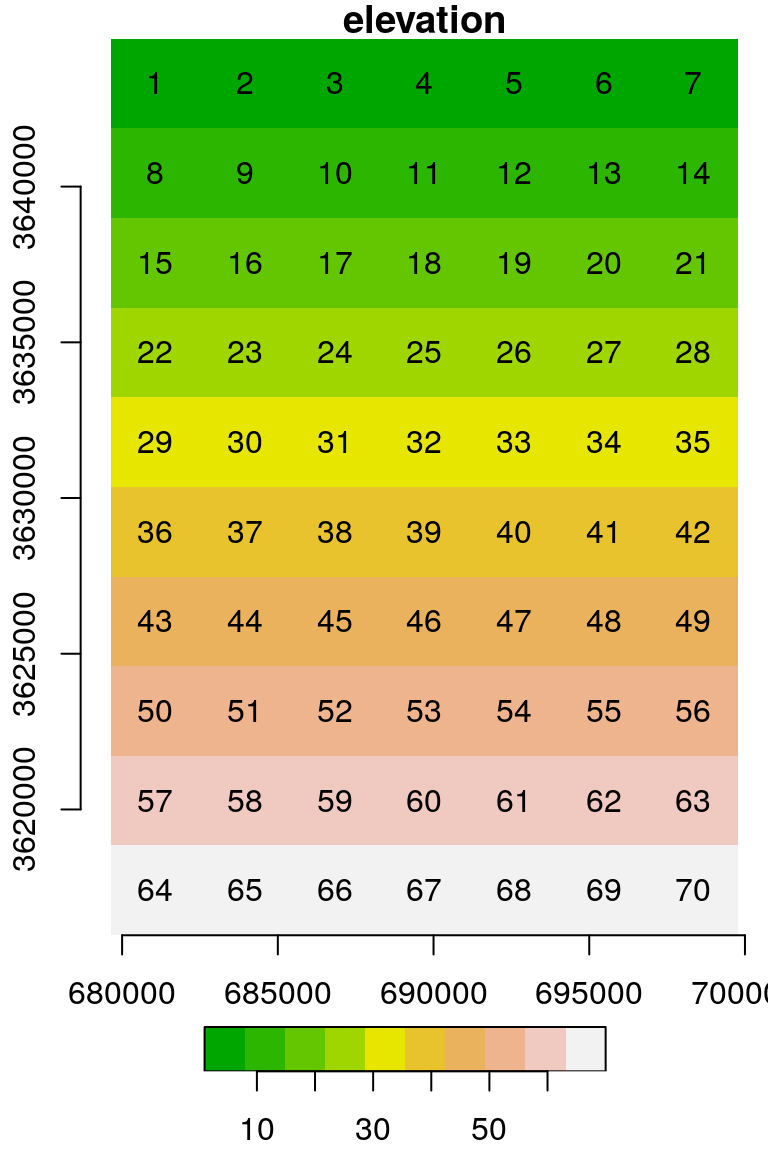
Figure 5.20: Raster with consecutive values
Sometimes it is useful to assign the same value to all cells, to create a uniform raster. This can be done using assignment of a single value, which is replicated, to the subset of all raster cells:
u = s
u[[1]][] = 1
u[[1]]
## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## [1,] 1 1 1 1 1 1 1 1 1 1
## [2,] 1 1 1 1 1 1 1 1 1 1
## [3,] 1 1 1 1 1 1 1 1 1 1
## [4,] 1 1 1 1 1 1 1 1 1 1
## [5,] 1 1 1 1 1 1 1 1 1 1
## [6,] 1 1 1 1 1 1 1 1 1 1
## [7,] 1 1 1 1 1 1 1 1 1 1The result is shown in Figure 5.21:
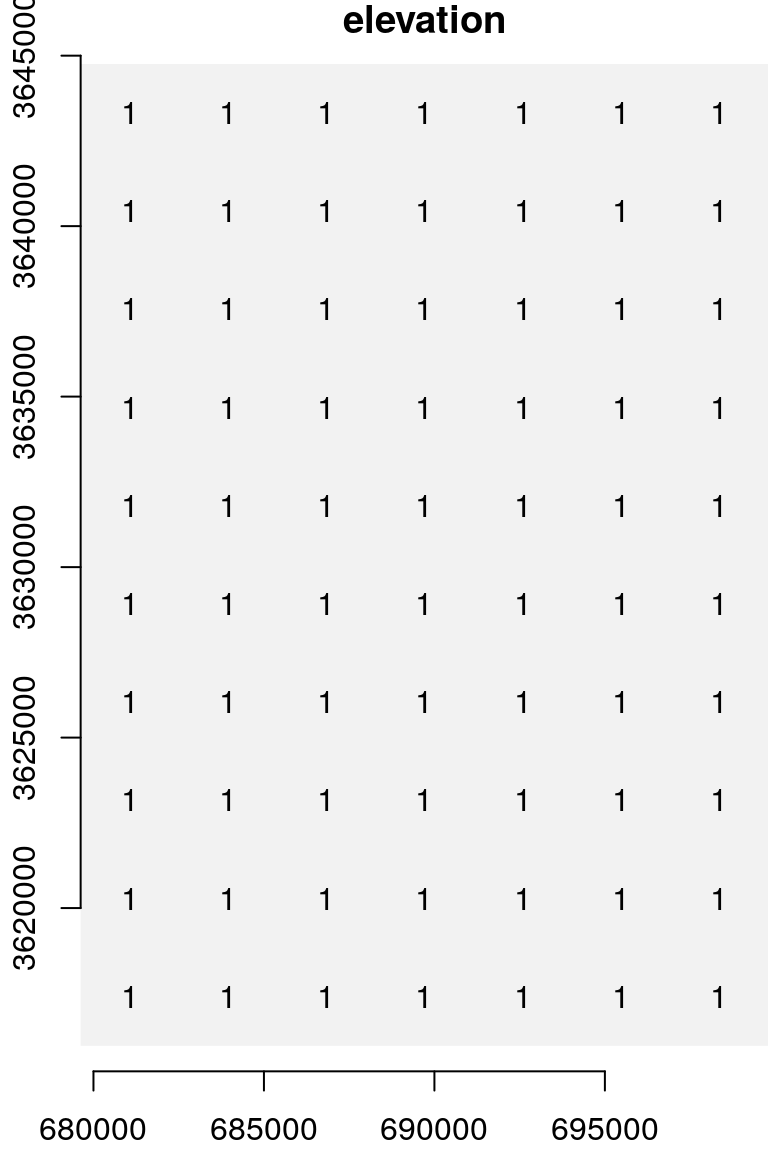
Figure 5.21: A uniform raster
5.3.9 The HDF format
HDF, which we mentioned above, is an example of a complex raster file format. Unlike a GeoTIFF file, an HDF file may contain more than one “attribute”, i.e., several matrices or arrays representing variables, as well as additional metadata, such as band names and units.
For example, the file named MOD13A3.A2000032.h20v05.006.2015138123528.hdf is an HDF file containing information from the MODIS satellite, namely the MOD13A3 product, for a particular date (2000-032) and a particular “tile” (h20v05):
Printing the resulting stars object reveals that the file contains 11 different attributes (each of them a \(1200\times1200\) matrix). The attributes indeed have informative names (such as "1 km monthly NDVI") and often also measurement units, such as degrees ([°]):
u
## stars object with 2 dimensions and 1 attribute
## attribute(s):
## elevation
## Min. :1
## 1st Qu.:1
## Median :1
## Mean :1
## 3rd Qu.:1
## Max. :1
## dimension(s):
## from to offset delta refsys point values
## x 1 7 679624 2880 UTM Zone 36, Northern Hem... FALSE NULL [x]
## y 1 10 3644759 -2880 UTM Zone 36, Northern Hem... FALSE NULL [y]5.3.10 Writing raster to file
We will not need to export rasters (or vector layers) in this book, since we will be working exclusively in the R environment. In practice, however, one often needs to export spatial objects from R to a file, to share them with other colleagues, further edit or process them in GIS software such as ArcGIS or QGIS, and so on.
Writing a stars raster object to a file on disk is done using write_stars. To run write_stars, we need to specify:
obj—Thestarsobject to writedsn—The file name to write
The function can automatically detect the required file format based on the file extension. For example, the following expression exports the stars object named s to a GeoTIFF file named dem_copy.tif in the current working directory:
There are similar functions named
rowSumsandcolSumsfor calculating row and column sums, respectively.↩Note that there are no data structure for zero-dimensional data structures (i.e., scalars) in R.↩
http://desktop.arcgis.com/en/arcmap/10.3/manage-data/raster-and-images/what-is-raster-data.htm↩
http://datacarpentry.org/organization-geospatial/01-intro-raster-data/index.html↩
http://matthewrocklin.com/blog/work/2018/02/06/hdf-in-the-cloud↩
R in a Nutshell, 2010.↩
These classes are both used to represent multi-band rasters. They differ only in their internal structure.↩
GeoTIFF files can come with both
*.tifand*.tifffile extension, so if one of them does not work you should try the other.↩